Biology Reference
In-Depth Information
both supported by 91% bootstrap values (Figure 4). Colombian Amazon haplotypes appear as
basal in this clade, and although Colombian Orinoco ones appear as a cluster, there is no
significant bootstrap support, and this cluster is also inconsistent using maximum parsimony
(data not shown).
BrA
BrC
BrD
BrB
BrE
BrF
BrA
BrC
BrD
BrB
BrE
BrF
CO5
CO1
CO3
CO2
CO4
CO5
CO1
CO3
CO2
CO4
51
51
98
98
91
91
96
96
CA1
CA2
BA6
BA2
BA4
BA1
BA5
BA3
BA7
CA1
CA2
BA6
BA2
BA4
BA1
BA5
BA3
BA7
91
91
53
53
Pontoporia_AY644451
Pontoporia_AY644451
0.05
0.05
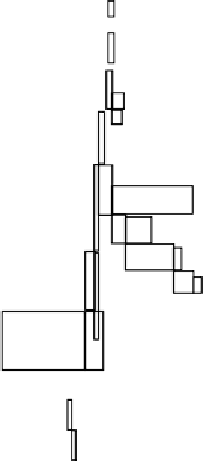
Figure 4. Neighbor-joining tree for HVSI mtDNA haplotypes. Bootstrap supporting values (percents of
1,000 replicates) are depicted on branches; only values above 50% are shown. CO (Colombian
Orinoco), CA (Colombian Amazon), BA (Bolivian Amazon) and Br (Brazilian Amazon) haplotypes.
We used a
Pontoporia blainvillei
control region sequence, retrieved from Genbank as an outgroup.
To address the systematic position of Brazilian
Inia
sp population, we included the 600
bp Cyt-b sequences reported by Banguera-Hinestroza et al., (2002). Phylogenetic
reconstruction of all
Inia
sp haplotypes shows that the Brazilian Amazon population
represents the most frequent haplotype discovered in the Colombian Amazon (CA2), in
agreement with the results obtained by these authors. However, it was not possible to
discriminate the two Brazilian haplotypes (Mamiraua-Tefe) in the tree, once the position of
the polymorphic site that separates them was not included in the 600 bp by Banguera-
Hinestroza et al., (2002) (data not shown).
The numbers of haplotypes (HT), polymorphic sites (S), mean pairwise difference (
d
),
gene diversity (
h
), nucleotide diversity (π) are displayed. Standard deviations are included for




















Search WWH ::

Custom Search