Biology Reference
In-Depth Information
remaining 5 genes belong to type III transcription factors, of which two resemble Sig F of
Synechocystis
sp. strain PCC 6803. Three others are distinctive from all other known Type III sigma factors.
xii) Genome of Oscillatoria
sp.
PCC 6506
:
The genome of
Oscillatoria
PCC 6506 is of 6.7 Mb.
Automatic annotation of the sequenced genome of
Oscillatoria
sp. PCC 6506 by MaGe annotation
software revealed 6,007 coding sequences among which genes encoding RNAs (84), photosynthesis
and
nif
gene cluster (37) and transposases (51) are recognized (Méjean
et al
., 2010b). Among the genes
encoding RNA, 70 genes have been allocated for tRNAs, 10 for small non-coding RNAs and 4 for
rRNAs. Overall 3.7% of the genes are responsible for the synthesis of secondary metabolites. The
gene clusters responsible for the synthesis of anatoxin-a and homoanatoxin-a (Cadel-Six
et al
., 2009;
Méjean
et al
., 2009, 2010a) and cylindrospermopsin (Mihali
et al
., 2008; Mazmouz
et al
., 2010) have
been identifi ed on the genome. In addition, four other gene clusters are dedicated for the synthesis of
polyketide synthase and non-ribosomal peptide synthetases. The presence of transposase sequences
close to the toxin encoding gene clusters denotes LGT events.
xiii) Genomes of C. raciborskii CS-505 and R. brookii D9
:
Both
C
.
raciborskii
CS-505 and
R
.
brookii
D9 constitute components of freshwater blooms while the strains of the former
produce cylindrospermopsin or paralytic shellfi sh poisons, the toxic strains of the latter produce
cylindrospermopsin and/or deoxycylindrospermopsin and anatoxin-a. The genomes of
C
.
raciborskii
CS-105 (3.89 Mb) and
R
.
brookii
D9 (3.2 Mb) are smaller in size amongst the fi lamentous and/or
fi lamentous, heterocystous forms comparable to the genome size of the unicellular
Synechocystis
sp.
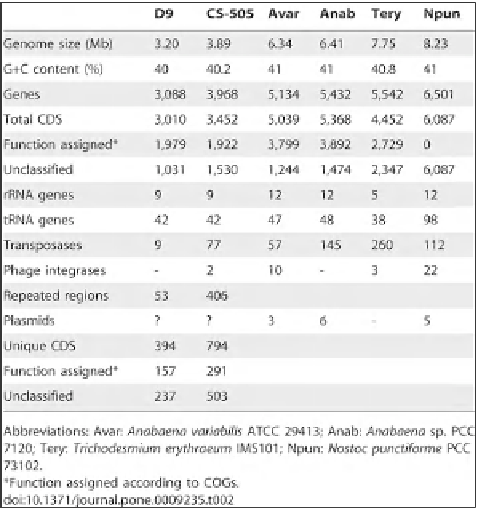
Table 4:
General features of the genomes of strains
Cylindrospermopsis raciborskii
CS-505 and
Raphidiopsis brookii
D9 in
comparison with four other fully sequenced genomes of fi lamentous cyanobacteria.
With the kind permission of
Mónica Vásquez
,
Department of Molecular Genetic and Microbiology, Faculty of Biological
Sciences, Pontifi cia Universidad Católica de Chile, Santiago, Chile & Millenium Nucleus EMBA, Santiago, Chile, [Stucken
et al
. (2010)
PLoS ON
E
5(2):
e9235. doi:10.1371/journal.pone.0009235].