Biology Reference
In-Depth Information
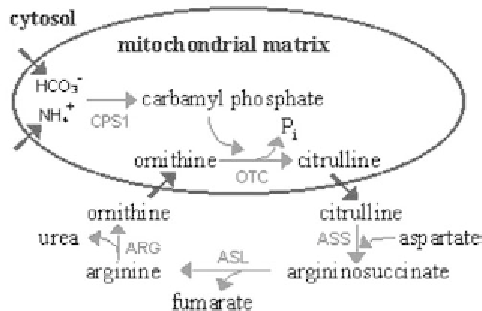
Fig. 6. Key enzymes in regulation of urea cycle in cells. CPS1: Carbamyl phosphate synthetase, EC 6.3.4.16; OTC: Ornithine
transcarbamylase, EC 2.1.3.3; ASS: Argininosuccinate synthetase, EC 6.3.4.5; ASL: Argininosuccinate lyase, EC 4.3.2.1;
ARG: Arginase, EC 3.5.3.1..
CASE STUDY
Inborn errors of metabolism
Inborn errors of metabolism are characterized by a block in a metabolic pathway, a deficiency of a
transport protein or a defect in a storage mechanism caused by a gene defect. The defect gene leads to
an absent or wrong production of essential proteins, especially enzymes that enable, disable or catalyze
the biochemical reactions of metabolic networks. Thus, these disorders of the metabolism result in a
threatening deficiency or accumulation of intermediate metabolites in the human organism and their
following corresponding symptoms.
For inborn errors of metabolism a lot of data is available in different databases accessible via the
Internet. Most inborn errors of metabolism are included in OMIM (Online Mendelian Inheritance
in Man
http://www3.ncbi.nlm.nih.gov/Omim
)
. Aside from the major molecular biological databases,
e.g., GenBank, EMBL, TRANSFAC, KEGG and BRENDA, our group's Metabolic Diseases Data
Base (MD-Cave) has been developed to simplify the collect and persistent storage of knowledge about
inborn metabolic diseases [Hofestaedt
et al.
, 2000b; Kauert
et al.
, 2001]. Although the amount of
this electronically available knowledge of genes, enzymes, metabolic pathways and metabolic diseases
increases rapidly, they give only highly specialized views of the biological systems. It is clearly that
the next task is to integrate all this knowledge and make it biotechnologically and medically applicable.
The MD-Cave is developed as such a bioinformatics system for representing, modeling and simulating
genetic effects on gene regulation and metabolic processes in human cells. In the following section, we
will present a case study that emphasizes on the modelling and simulation of the gene regulated urea
cycle network by using the hybrid Petri net.
Urea Cycle and its regulation
In human cells, excess nitrogen is removed either by excretion of NH
4
(of which only a little happens)
or by excretion of urea. Urea is largely produced in the liver by the urea cycle, a series of biochemical
reactions that are distributed between the mitochondrial matrix and the cytosol (Fig. 6). The cycle centers
around the formation of carbamyl phosphate in hepatocyte mitochondria to pick up NH
4
and incorporate