Biology Reference
In-Depth Information
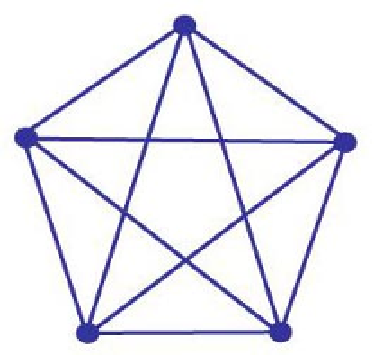
Fig. 10.3.
A clique of size five.
The
paraclique
algorithm was recently introduced in [6], where it was shown
to have advantages in the amelioration of noise inherent in high throughput bio-
logical data. Clique by itself is highly resistant to false positives. Under certain
experimental conditions, however, it can be subject to false negatives. This is
because, if even a single edge is missing, the clique is lost. Moreover, we fre-
quently encounter enormous numbers of overlapping cliques [19]. To coalesce
these into fewer but larger clusters, and to reduce the significance of noise, par-
aclique solves something similar to the dense-k-subgraph problem [10], which is
NP
-complete even on graphs of maximum degree three. Roughly speaking, a
paraclique is a clique augmented with non-clique vertices in a highly controlled
manner. A user-defined
glom factor
,
g
, is provided to increase cluster size while
limiting the number of missing edges permitted. We glom onto a non-clique ver-
tex only if it is adjacent to at least
g
clique/paraclique members. This notion is
depicted in Fig. 10.4. Correlations between non-adjacent vertices may be taken
into consideration as well. We refer the reader to [6] for details. Thus, when the
application permits, we employ the paraclique algorithm and sacrifice overlap in
order to build robust clusters.
Paraclique is also useful from a computational standpoint because it can, de-
pending on the application, obviate the need for maximal clique enumeration. To
illustrate, the processing of an NOD file whose maximum clique size was only 20
produced a list containing over four million maximal cliques and requiring over
two gigabytes of memory before the enumeration was terminated by the operating
system. In contrast, only 25 paracliques were generated. We therefore identify a
maximum clique, use paraclique to decompose the graph, and then iterate the pro-
cess on the remaining subgraph. We halt the process when maximum clique size







Search WWH ::

Custom Search