Biology Reference
In-Depth Information
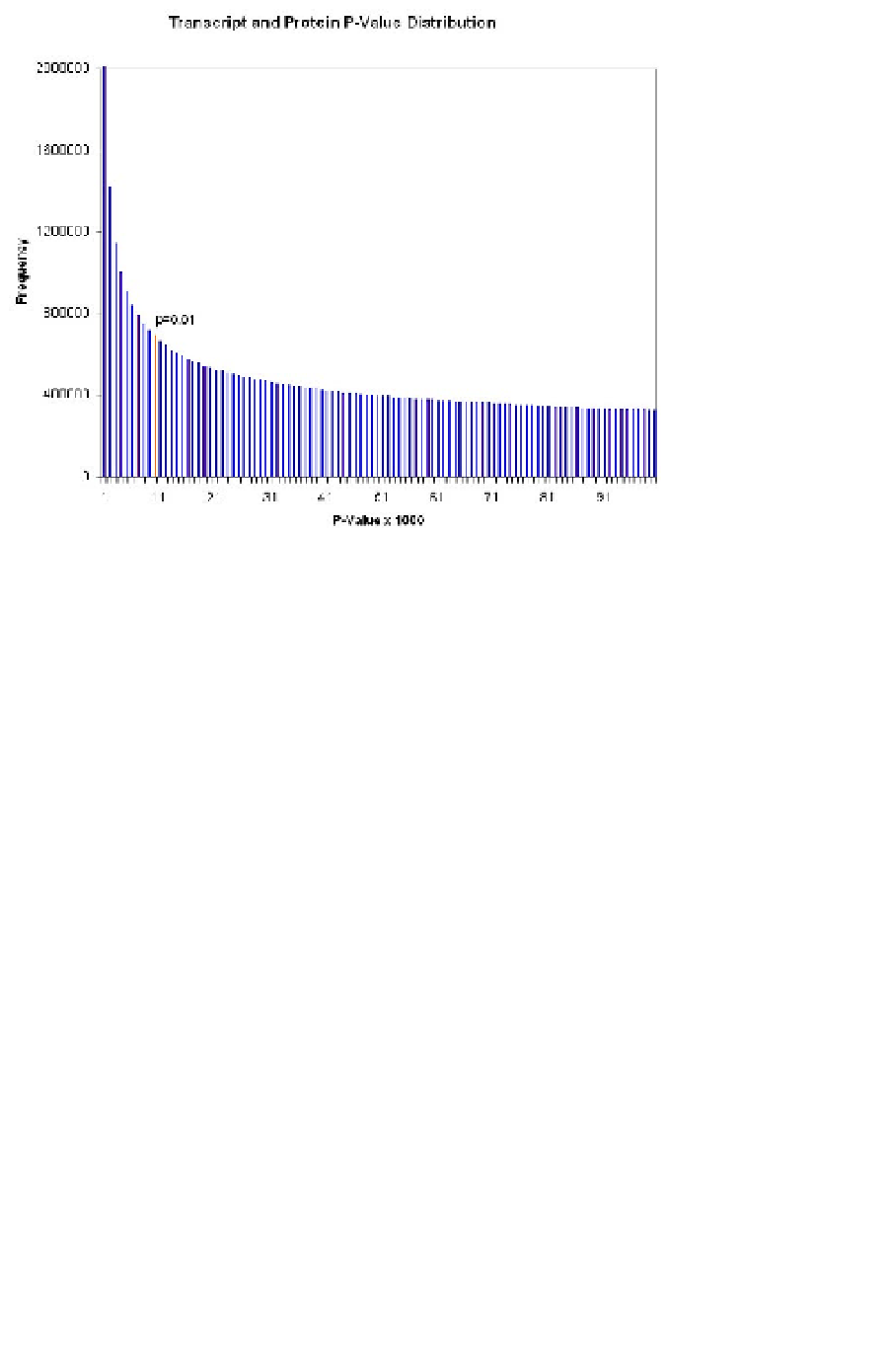
Fig. 10.2. The transcriptomic data used in this study provides a broad spectrum of p-values. A
threshold p-value of 0.01, for example, creates an unweighted graph with 22750 vertices and roughly
11 million edges.
ular biology [23]. In the present setting, its advantages include cluster purity (all
edges are present), cluster overlap (genes and gene products are pleiotropic), and
resistance to false positives (the bane of many clustering methods). Contrasts with
other techniques can be found in [28]. The classic
decision
version of clique is
NP
-complete. Finding approximate solutions appears no easier, because ensur-
ing solutions within
n
in polynomial time implies
for any
>
0 [9].
We are of course more interested in
search
and
optimization
. By transform-
ing clique to vertex cover, we can apply notions from fixed-parameter tractabil-
ity [1, 8] and many years of basic research [11, 12] to solve the
maximum
clique
problem effectively in practice. With novel implementations and high perfor-
mance platforms, we are currently able to find maximum cliques with hundreds
of vertices in graphs with tens of thousands of nodes. We must often also solve
the
maximal
clique problem [5]. Even when the maximum clique size is mod-
est, we frequently find that the number of maximal cliques is staggering. Thus it
is that space as well as time is a critical resource for solving maximal clique,
even when supercomputing technologies are used. Our work on this general
subject, as well as its application to transcriptomic data analysis, is chronicled
in [1, 4, 6, 7, 19, 20, 28, 34].
P
=
NP







Search WWH ::

Custom Search