Biomedical Engineering Reference
In-Depth Information
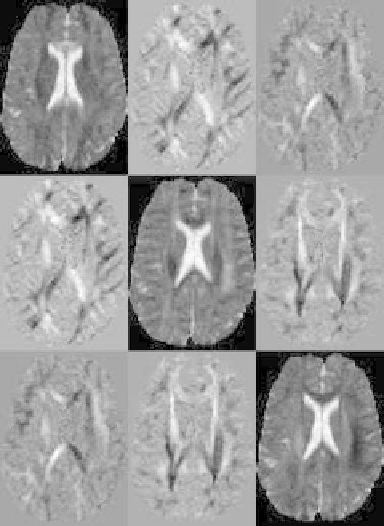
Figure 8.11: Slice of a tensor volume where every “element” of the image matrix
corresponds to one component of the tensor D.
orientations of the sample or field gradient and therefore cannot themselves be
used for classification purposes. Moreover, 3D visualization and segmentation
techniques available today are predominantly designed for scalar and sometimes
vector fields. Thus, there are three fundamental problems in tensor imaging: (a)
finding an invariant representation of a tensor that is independent of a frame of
reference, (b) constructing a mapping from the tensor field to a scalar or vector
field, and (c) visualization and classification of tissue using the derived scalar
fields.
The traditional approaches to diffusion tensor imaging involve converting
the tensors into an eigenvalue/eigenvector representation, which is rotationally
invariant. Every tensor may then be interpreted as an ellipsoid with principal
axes oriented along the eigenvectors and radii equal to the corresponding eigen-
values. This ellipsoid describes the probabilistic distribution of a water molecule
after a fixed diffusion time.
Using eigenvalues/eigenvectors, one can compute different anisotropy mea-
sures [55, 57-59] that map tensor data onto scalars and can be used for further