Biology Reference
In-Depth Information
a
b
M
(kDa)
XS
XSConA
170
130
95
72
55
protein sample
M
(kDa)
43
1
72
34
34
26
15
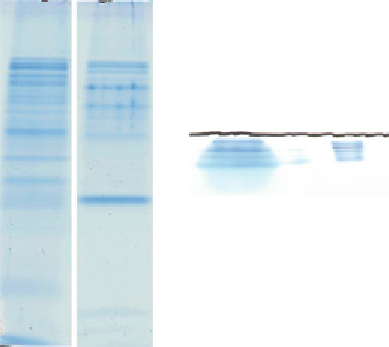
Fig. 2
Separation of proteins by SDS-PAGE prior to MS analysis. (
a
) A long run is
performed prior to sampling for MALDI-TOF MS analysis. XS stands for xylem sap
proteins and XSConA for xylem sap proteins eluted from a ConA affi nity column.
The arrow points at ConA leaking from the column. Each stained protein band is
cut out of the polyacrylamide-SDS gel and prepared for MS analysis. (
b
) A short
run is performed prior to sampling for LC-MS/MS analysis. The thick stained
band is cut in three parts delimited by dotted lines (1-3) and each of them is
prepared for MS analysis. The polyacrylamide gels are stained with colloidal
blue. Molecular mass markers (M) are indicated
3. Combine fi rst and second elution fractions. Desalt using
Econo-Pac 10 DG columns and lyophilize as described
(
see
Subheading
3.3
,
step 2
). The sample obtained is the
“xylem sap
N
-glycoproteome.”
1. Resuspend the two dried protein samples (“xylem sap pro-
teome” and “xylem sap
N
-glycoproteome”) in 300
3.5 Separation of
Proteins by SDS-PAGE
μ
L and
100
L of each sample
on a 10 × 12 × 0.15 cm SDS-polyacrylamide gel with stacking
gel and resolving gel at a concentration of 4 %/0.11 % and
12.50 %/0.33 % of acrylamide/bisacrylamide respectively.
μ
L of UHQ water respectively. Load 50
μ
2. For MALDI-TOF (Matrix-Assisted Laser Desorption
Ionization-Time Of Flight) MS analyses, perform separation
by SDS-PAGE at 20 mA per gel for the stacking gel and at
40 mA per gel for the resolving gel, until the front line reaches
the bottom of the gel (total migration time of about 75 min)
(long run, Fig.
2a
).
3. For LC (Liquid Chromatography)-MS/MS analyses, perform
separation by SDS-PAGE at 20 mA per gel for the stacking and
the resolving gels until the proteins migrate about 6 mm in