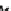Biology Reference
In-Depth Information
harvesting
xylem sap
dialysis
dialyzed xylem sap
ConA affinity
chromatography
desalting
lyophilization
desalting
lyophilization
xylem sap proteome
xylem sap
N
-glycoproteome
SDS-PAGE separation
long run
short run
MALDI-TOF MS
LC-MS/MS
xylem sap protein data / bioinformatic analyses:
identification, sub-cellular localization,
functional domains,
N
-glycosylation
Fig. 1
Strategy for xylem sap proteomic analysis from harvesting of xylem sap to
protein identifi cation by mass spectrometry and bioinformatics analysis
2. Desalt half of this dialyzed sample using Econo-Pac 10 DG
columns to get the “xylem sap proteome.” Equilibrate the
columns with 20 mL of buffer B2. Load 3 mL of dialyzed
xylem sap sample onto each column. Elute proteins with 4 mL
of buffer B2. Pool elution fractions and lyophilize resulting
desalted sample overnight.
3. Quantify proteins by the Bradford method following the
Uptima Coo Protein assay Kit (
see
Note 14
).
4. Use the second half of the dialyzed sample to get the “xylem
sap
N
-glycoproteome” (
see
Subheading
3.4
.)
3.4 Affi nity
Chromatography on
ConA ( See Note 15 )
1. Perform ConA chromatography in batch mode to capture
xylem sap
N
-glycoproteins (
see
Note 16
).
2. Prewash the resin (0.6 mL) with 20-fold volumes of buffer B3
(
see
Note 17
) and equilibrate with tenfold volume of buffer B1.
Mix the dialyzed xylem sap sample (10 mL) with the matrix in
batch for 1 h at 4 °C under mild shaking. Wash the resin three
times with 1.5 mL of buffer B1 after fl ow-through removal.
Elute proteins with 1.5 mL of buffer B4. Repeat elution twice.