Biomedical Engineering Reference
In-Depth Information
o
2
, …, o
j
, called “observed frequencies”, and that,
according to the rules of probability, they would
be expected to occur with frequencies e
1
, e
2
, …,
e
j
, called theoretical or expected frequencies, as
shown in Table 3.
A measure of the discrepancy between the
observed and expected frequencies is given by
the
level. Otherwise, the H
0
will be accepted or,
at
least, not rejected.
Now, looking at Table 4 taken from Jack Lester
King and Thomas H. Jukes (1969), we ran the
2
test on these data and found that the value of
2
is 477.809. As this value is much greater than
the expected
2
0
, which, for the 19 degrees of
freedom in respect of the twenty amino acids, is
38.6, we find that the observed frequencies dif-
fer very significantly from the expected values,
thereby rejecting the H
0
. Accordingly, it is to
be expected that the bases are not associated as
triplets at random and, therefore, an explanation
needs to be sought.
95
2
statistic, as
(
)
(
)
(
) (
)
2
2
2
2
o
−
e
o
−
e
o
−
e
o
−
e
j
∑
=
j
j
j
j
2
=
1
1
+
2
2
+
+
=
e
e
e
e
i
1
1
2
j
j
where, if the total frequency is N,
.
∑
o
=
∑
e
=
N
i
j
If
2
= 0, the observed and expected frequen-
cies are exactly equal, whereas if
2
> 0, they are
not. The greater the value of
2
, the greater the
discrepancies between the two frequencies are.
The
DECIPHERING THE GENOME
2
sample distribution is very closely
approximated to the chi-square distribution
given by
As already mentioned, genes are usually located
in the chromosomes: cellular structures whose
main component is deoxyribonucleic acid, ab-
breviated to DNA. DNA is formed by comple-
mentary chains, made up of long sequences of
nucleotide units. Each nucleotide contains one
of the four possible nitrogenised bases: adenine
(A), cytosine (C), thymine (T) and guanine (G),
which only associate in two possible ways with
each other: A with T and C with G. Usually, some
portions of DNA form genes and others do not.
In the case of human beings, the portions that are
genes make up only approximately 10% of total
DNA. The remainder appears to have nothing
to do with protein synthesis; it is, until it finds
a functionality, genetic trash so to speak. In any
case, the reading and interpretation of this set of
symbols that make up DNA can be compared to
deciphering the hieroglyphics of life. If Jean-Fran-
çois Champollion's deciphering of hieroglyphic
script from the Rosetta Stone was arduous and
difficult, imagine deciphering 3x10
9
symbols
from a four-letter alphabet. To give an idea of the
magnitude of the endeavour, suffice it to say, for
example, that the sequence of DNA nucleotides
written would take up a space equivalent to 150
( )
(
1
1
)
1
−
Y
=
Y
2
v
−
2
e
2
=
Y
v
−
2
e
2
2
2
2
0
0
where ν is the number of degrees of freedom
given by:
a. ν = k-1, if the expected frequencies can
be calculated without having to estimate
population parameters from the sample
statistics.
b. ν = k-1-m, if the expected frequencies can
only be calculated by estimating m param-
eters of the population from the sample
statistics.
In practice, the expected frequencies are
calculated according to a null hypothesis H
0
. If,
according to this hypothesis, the calculated value
of
2
is greater than any critical value, such as
2
2
0
, which are the critical values at the
significance levels of 0.05 and 0.01, respectively,
it is deduced that the observed frequencies differ
significantly from the expected frequencies and
the H
0
is rejected at the respective significance
or
0
95
99
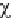

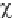

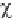
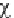







Search WWH ::

Custom Search