1. Introduction
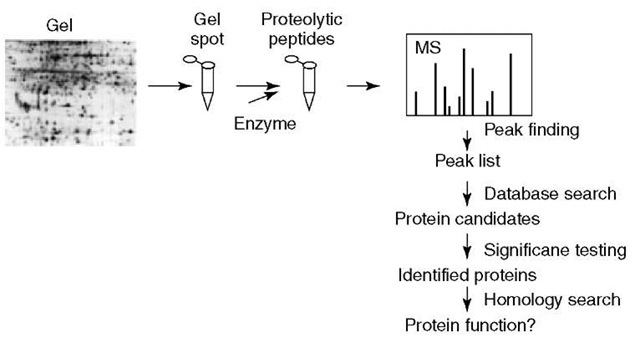
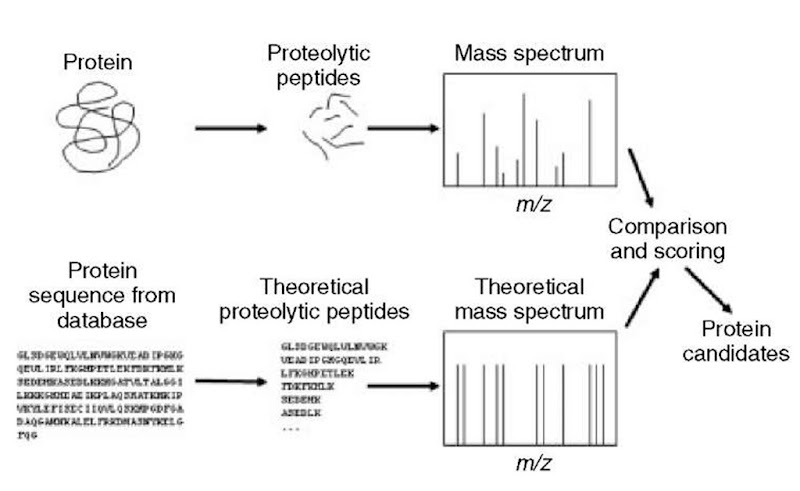
Identification of proteins by searching protein sequence collections with peptide mass fingerprinting data is widely used (Figure 1). The proteins in the sample are first separated to obtain one or a few proteins of interest. These proteins are then digested with a proteolytic enzyme. Mass spectra of the resulting peptide mixtures are acquired. The mass spectra are processed to find the masses of the peptides in the mixture. These measured masses are compared with calculated peptide masses for each protein in a protein sequence collection according to the rules defined by a set of user-defined parameters (Figure 2). A score is calculated for each comparison and the protein sequences in the collection are ranked according to the calculated score. Different search engines calculate the score in different ways (Henzel et al., 1993; Mann et al., 1993; Pappin et al., 1993; Yates et al., 1993; James et al., 1993; James et al., 1994; Wilkins et al., 1998; Perkins et al., 1999; Clauser et al., 1999; Berndt etal., 1999; Gras etal., 1999; Zhang and Chait, 2000; Gay etal., 2002; Eriksson and Fenyo, 2004a; Samuelsson etal., 2004; Rognvaldsson etal., 2004; Magnin etal., 2004; Levander etal., 2004). The meaning of these scores is in general not easily understood by the nonexpert user and they are not amenable to automation. Therefore, an additional step is necessary to test the significance of the results and convert the search engine-dependent score into a measurement of the significance of the protein identification result.
The problem of comparing the experimental mass spectra with calculated peptide masses for a protein sequence collection has been solved with different approaches in the different search engines. However, all search engines calculate a score for ranking the proteins in the sequence collection. The best matching protein sequence has the highest probability of being present in the sample that was analyzed and it is therefore ranked highest. The most widely used search engines are Mascot (Perkins etal., 1999), ProFound (Zhang and Chait, 2000), and MS-Fit (Clauser etal., 1999). For an evaluation of the performance of the different peptide mass fingerprinting algorithms, see Chamrad et al. (2004).
This paper will discuss how different search parameters influence the results and how the search engine-dependent scores can be converted into a measurement of the significance of the protein identification result.
Figure 1 Protein fingerprinting is typically performed by first separating the proteins followed by enzymatic digestion and mass analysis. The raw mass spectra are subsequently analyzed to obtain a list of peptide masses. This peptide mass map is then searched against a protein sequence collection and the significance of each protein candidate is tested
Figure 2 Searching a sequence collection with peptide mass fingerprinting data is performed by mimicking the experiment in silico: each entry in a protein sequence collection is theoretically digested using the same specificity as the enzyme used in the experiment. A theoretical mass spectrum is constructed, compared with the measured mass spectrum. The entries in the protein sequence collection are ranked according to how well they match the experimental data
2. Significance testing
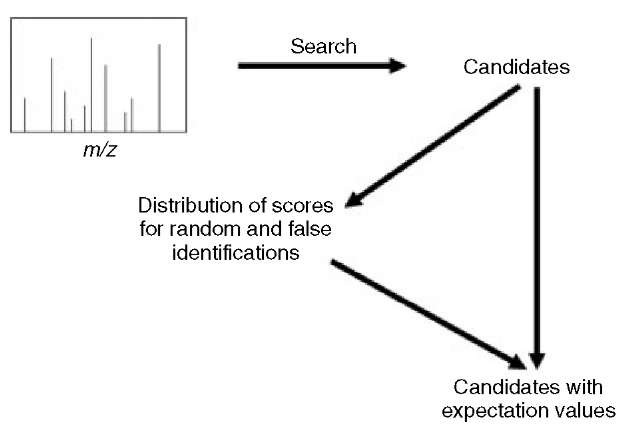
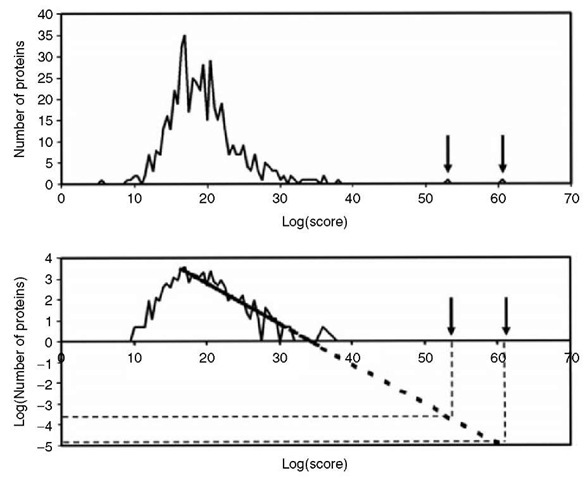
One of the critical steps in protein identification is significance testing (Eriksson and Fenyo, 2004a; Eriksson et al., 2000; Eriksson and Fenyo, 2002; Fenyo and Beavis, 2003; Eriksson and Fenyo, 2004b). False identifications are possible due to random matching between the measured and calculated masses. In the result of a search of a sequence collection with peptide mass fingerprinting data, there will always be a highest ranked protein sequence. This protein sequence might correspond to a protein that is in the sample analyzed or simply be a false-positive, that is, get the highest score because of random matching between the calculated and measured proteolytic peptide masses. The probability that a protein candidate is a false-positive can be estimated by comparing its scores to the distribution of scores for random and false identifications. The distribution of scores for random and false identifications can be obtained by collecting statistics during the search. Figure 3 illustrates the method for using the statistics collected during the search to estimate the significance of the results. A score is calculated for each protein sequence in the collection. For the majority of sequences, the matching with the experimental data is random. An example of the distribution of the scores for proteins in a sequence collection matching a peptide mass fingerprint is shown in Figure 4. Typically, a distribution of scores from randomly matching protein sequences is observed at low scores. This distribution is an extreme value distribution, having a linear tail when plotted on a log-log scale. The expectation value of high-scoring protein sequences is estimated by extrapolation.
Figure 3 Most protein candidates given by a search engine are due to random matching. The distribution of scores for random and false identifications can therefore be obtained and used to calculate the expectation value for the protein candidates
3. Search parameters
The different search engines available for peptide mass fingerprinting have a similar set of parameters, including enzyme specificity, sequence collection, modifications, peptide mass tolerance, protein mass, and pi. In cases in which the data quality is very high, a significant result will be obtained with the search parameters set within a wide range. However, in most cases, it is critical to select the parameters carefully when searching a protein sequence collection with peptide mass fingerprinting data. In general, it is recommended that all the information available that can restrict the search be used, for example, if the origin of the sample is known, a lot can be gained by only searching the species of interest and not all known protein sequences from all organisms. Also, information that will increase the number of possible matching peptides in the dataset should be used conservatively, for example, partial modification such as phosphorylation.
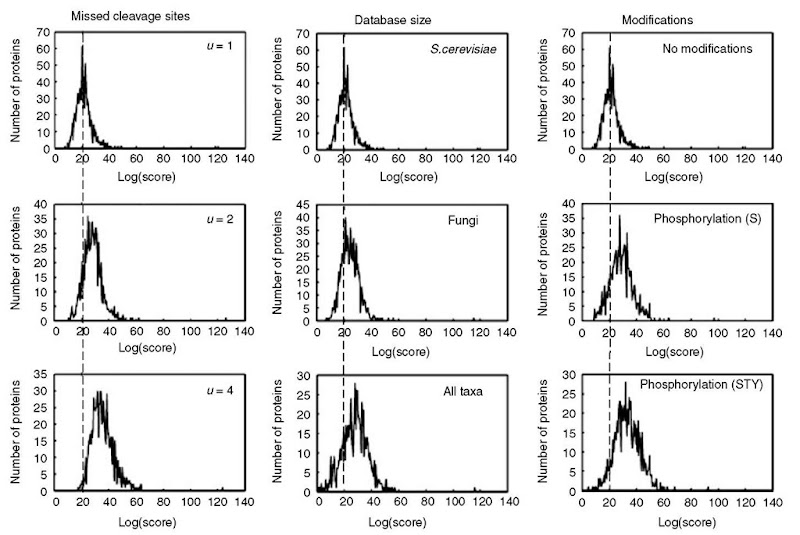
It is important to use enzymes with high specificity for peptide mass fingerprinting. The number of incomplete cleavage sites selected will influence the results; a larger number of incompletes will increase the noise because there will be more possible peptide sequences. This is illustrated in Figure 5 in which the distribution of scores for the random matching shifts to higher scores when the number of possible missed cleavage sites allowed is increased from 1 to 2 and 4. It is therefore recommended that an experimental protocol be used in which the proteins are digested as completely as possible, allowing the search to be performed with a setting of 0 or 1 incomplete cleavage sites.
Figure 4 An example of the distribution of scores for random and false identifications for a peptide mass fingerprinting search with ProFound (Zhang and Chait, 2000). The distribution is an extreme value distribution, having a linear tail when plotted on a log-log scale. The expectation value of high-scoring protein sequences is estimated by extrapolation
There are many different sequence collections available for searching. Protein sequence collections are the most common choice for protein identification with peptide mass fingerprinting data. Searching of raw genomic data by translating the entire DNA sequence in all six possible reading frames with peptide mass fingerprinting data can be successful for organisms with small genomes, but is generally not done, because it requires very high quality experimental data. Expressed sequence tag (EST) collections are in general unsuited for searching with peptide mass data, because of their incomplete coverage of the genes. The smaller the sequence collection searched, the higher the significance of the results, provided that the collection contains the sequence of interest (Figure 5), for example, if the origin of the sample is known, it is recommended that only the species of interest be searched.
Figure 5 Example showing how the distribution of scores of random matches changes when different parameters are used with the same data set in searches with ProFound.
Table 1 Example illustrating how the expectation value changes when different parameters (size of sequence collection, missed cleavage sites – u, and modifications) are used with the same data set in searches with ProFound.
| Sequence collection | E-value | u | E-value | Modifications | E-value |
| S. cerevisiae | 4.8E-07 | 1 | 4.8E-07 | No modifications | 4.8E-07 |
| Fungi | 8.4E-06 | 2 | 1.1E-05 | Phosphorylation (S) | 2.3E-03 |
| All taxa | 2.9E-04 | 4 | 6.8E-04 | Phosphorylation (STY) | 2.1E-02 |
Proteins are often naturally modified and usually deliberately or unintentionally modified in the sample preparation process. Modifications can be defined as complete (i.e., they are always present on a specific amino acid) or partial (i.e., the modification might or might not be present on an amino acid). Complete modifications (e.g., cystein alkylation) do not increase the search time or change the significance. In contrast, partial modifications (e.g., phosphorylation) increases the search time and in general decreases the significance (Table 1) because the distribution of scores for the random matching shifts to higher scores (see Figure 5). If a peptide contains an amino acid that potentially might be modified, the mass of the unmodified peptide and the masses of the peptide with all possible modifications have to be compared with the measured peptide mass map. Even though a lot of proteins are modified, most proteins have only a few modified amino acids and therefore only a few of the peptides in a peptide mass map will be modified. Searching with partial modifications defined will increase the random background and potentially the number of matching peptides. In most cases, higher significance is achieved when searching without partial modifications defined.
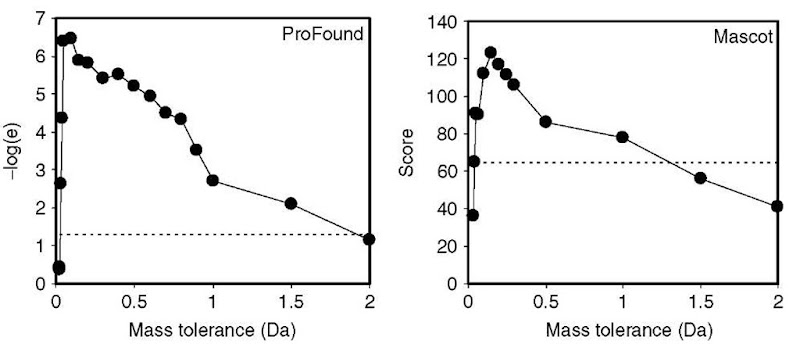
The quality of the results obtained is dependent on the mass tolerance selected (Figure 6). Increasing the mass tolerance will increase the number of both true-and false-matching peptides. The two extreme cases are (1) zero mass tolerance -no peptides match and (2) large mass tolerance – all peptides match. Therefore, the best results are obtained at the mass tolerance that balances the contributions from true- and false-matching peptides. The best setting for the mass accuracy can differ for different search engines because of the differences in the algorithms, for example, in Figure 6, the best results are obtained with a mass tolerance of 0.15 for Mascot and 0.1 for ProFound.
Figure 6 An example showing how the mass tolerance affects the results of the search for ProFound (Zhang and Chait, 2000) and Mascot (Perkins et al., 1999). The dotted line shows the probability of 0.05 for the result being false
If the source of the analyte is a spot in 2D gel, information on protein properties such as mass and pI is available. This information can be used to restrict the search by excluding all sequences in the collection being searched that do not match the measured mass and pI and thereby increasing the significance of the results. The tolerance for protein mass and pI should, however, not be set too narrow because the measurement can differ from the calculated because of several reasons: (1) the protein has been processed and only a small domain is observed; (2) the splice variant observed is not in the sequence collection; and (3) the intron-exon boundaries are incorrectly assigned.
4. Summary
For successful protein identification, it is necessary to (1) use a sensitive and selective algorithm; (2) carefully select search parameters; and (3) test the significance of the results. The potential for obtaining a true mass spectrometric protein identification result depends on the choice of algorithm as well as on experimental factors that influence the information content in the mass spectrometric data. Current methods can never definitely prove that a result is true, but an appropriate choice of algorithm can provide a measure of the statistical risk that a result is false – that is, the statistical significance – and guide the practitioner in interpreting the results.