INTRODUCTION
Bioinformatics involves the design and development of advanced algorithms and computational platforms to solve problems in biomedicine (Jones & Pevzner, 2004). It also deals with methods for acquiring, storing, retrieving and analysing biological data obtained by querying biological databases or provided by experiments. Bioinformatics applications involve different datasets as well as different software tools and algorithms. Such applications need semantic models for basic software components and need advanced scientific portal services able to aggregate such different components and to hide their details and complexity from the final user. For instance, proteomics applications involve datasets, either produced by experiments or available as public databases, as well as a huge number of different software tools and algorithms. To use such applications it is required to know both biological issues related to data generation and results interpretation and informatics requirements related to data analysis.
Bioinformatics applications require platforms that are computationally out of standard. Applications are indeed (1) naturally distributed, due to the high number of involved datasets; (2) require high computing power, due to the large size of datasets and the complexity of basic computations; (3) access heterogeneous data both in format and structure; and finally (5) require reliability and security. For instance, applications such as identification of proteins from spectra data (de Hoffmann & Stroobant, 2002), querying of protein databases (Swiss-Prot), predictions of proteins structures (Guerra & Istrail, 2003), and string-based pattern extraction from large biological sequences, are some examples of computationally expensive applications. Moreover, expertise is required in choosing the most appropriate tools. For instance, protein structure prediction depends on proteins family, so choosing the right tool may strongly influence the experimental results.
Recently, there has been much interest from database community and computer science community for bioinfor-matics. Nevertheless, what is still missing is a high-level environment able to classify tools and provide Web-based easy to use application programming interfaces. In such a way, users can concentrate on the logic of application (i.e., biological aspects) leaving to such platform the work to compose applications, format input data, provide options and parameters, and collect results.
Another important requirement is the accessibility of such platform through a Web portal, that is, by using the user interfaces and protocols of the World Wide Web. A bioinformatics Web portal is thus a Web portal that allows access to bioinformatics tools and databases through a Web browser. Moreover, due to the complexity, diversity and a huge number ofbioinformatics tools and databases, a bioinformatics Web portal should also support problem formulation, application composition and execution, results visualisation and annotation. A possible approach to solve these issues —high-level modeling and Web-based user interfaces—can be obtained by adding semantics links between biological problems and bioinformatics resources through ontologies (Baker, 1998), and by decoupling Web-based user interfaces from high-performance back-end platforms.
In this article we review main requirements of distributed bioinformatics applications and related bioinformatics Web portals, and report the proposal of a grid-based bioinformatics portal allowing choosing and composing of bioinformatics tools with the help of a domain ontology describing data and software resources.
BACKGROUND
Bioinformatics researchers, among the other directions, are investigating through: (1) data modeling to manage heterogeneous datasets (e.g., see HUPO, n.d., the HUPO, Human Proteome Organization—Proteomics Standard Initiative); (2) specialised services for protein sequences searching, and data mining techniques to extract meaningful information from datasets; (3) ontologies and metadata for a high-level description of the goals and requirements of applications; and (5) high performance computational platforms to execute distributed bioinformatics applications.
Many applications have been defined to support biological researchers for solving problems on different topics where large computing power is required. Grid community (Foster & Kesselman, 2003) has recognised that bioinformatics and postgenomic applications are both a challenge but especially an opportunity for distributed high performance computing and collaboration. The Life Science Grid Research Group of the Global Grid Forum (see LSG, n.d.) aims to investigate how bioinformatics requirements can be fitted and satisfied by grid services and standards, and vice versa, what new services should grids provide to bioinformatics applications. Some bioinformatics grids projects are also appearing, for example, the EuroGrid project (EuroGrid, n.d.), the Bio-GRID work package (Bio-Grid, n.d.) used to access portal for biomolecular modeling resources, the myGrid (Stevens, Robinson, & Goble, 2003) system, and the Asia Pacific Grid (AsiaGrid, n.d.).
In recent years many platforms for developing bioinformatics applications, some of which dealing with ontologies and workflows, have been developed. Systems as SpecAlign (Wong, Cagney, & Cartwright, 2005), MSAnalyzer (Sashimi, n.d.), and those developed in Jeffries (2005), are all specialised in preprocessing, visualisation, and analysis of specialised datasets, that is, mass spectrometry data, but they do not support analysis of data and workflows composition, nor include domain ontologies. LabBase (Goodman, 1998) and similar laboratory information management systems are useful to manage experiments conducted in laboratory and related data, but are inadequate to support sophisticated analysis. More sophisticated bioinformatics platforms, like the genomics research network architecture (gRNA) (Laud, Bhowmick, Cruz, Singh, & Rajesh, 2002) and the Pegasys (Shah et al., 2004) bioinformatics system, offer some sort of configurable engine to pipeline a set of tasks and data. A special attention merits myGrid (Stevens et al., 2003), a powerful toolkit to build workflows of Web services that offers a large set of bioinformatics tools wrapped as Web services, leverages ontologies, and uses the powerful Taverna workflow editor (Oinn et al., 2004). General purpose workflow editors (see Yu & Buyya, 2005, for a survey), such as Kepler, Pegasus, and Triana, are all suitable to support the composition of bioinformatics workflows, but few of them use ontologies.
Finally, some bioinformatics Web portals are also appearing. Such systems, some of which are described in the following, offer a collection of bioinformatics tools and provide access to local and remote biological databases through a Web-based interface, but a few of them offer a machine-understandable semantic classification of the tools nor gives support for the design of complex workflows of such tools. The ExPASy (n.d.; Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics is dedicated to the analysis of protein sequences and structures (ExPASy). The grid protein sequence analysis (GPSA) is an integrated grid portal devoted to molecular bioinformatics and offers a user-friendly interface for the grid genomic resources on the EGEE grid. The Helmholtz Network for Bioinformatics (HNB, n.d.) offers access to numerous bioinformatics resources provided by many Germanbioinformatics research groups through a single Web portal. Mobyle (Neron, Tuffery, & Letondal, 2005), is an environment for running and defining bioinformatics analyses whose main objective is to enable biologists to access advanced features, such as pipelines or remote services discovery, without having to learn complex concepts nor installing sophisticated software.
In summary, an important trend in bioinformatics environments regards the increasing use of ontologies to model basic building blocks and the use of workflow systems to ease the application development and execution process in a distributed setting such as the grid. The decoupling between user interface and execution back-end is another important trend to move such environments toward bioinformatics Web portals.
REQUIREMENTS OF BIOINFORMATICS WEB PORTALS
From a computational point of view, bioinformatics applications present the following requirements:
1. They are often distributed, due to the high number of involved datasets.
2. They require high computing power, due to the large size of datasets and the complexity of basic computations.
3. They access heterogeneous data, where heterogeneity is in data format, access policy, distribution, and so forth.
4. They could access private data, thus should be based on a secure software infrastructure.
Current biological and biomedical research, for example, genomics and proteomics, makes full use of a plethora of tools and databases that address specific problems such as nucleotide/protein sequence alignment (e.g., see BLAST, n.d.), protein structure prediction, protein docking, mass spectrometry-based protein identification (e.g., see MASCOT, n.d.), molecule visualisation (e.g., see RasMol, n.d.), and so forth. Although many of those tools and databases are made available on the Internet, often researchers use them in a stand-alone way and if an experiment needs a composition of such tools, users need to manually insert input data and collect output results that in turn are used to feed another tool.
Current bioinformatics Web portals are just a collection of those tools and the more sophisticated provide also access to remote databases, but they do not offer support for the design and execution of complex “in silico” experiments.1 Thus, next-generation bioinformatics Web portals need to support the entire lifecycle of in silico experiments, that is:
1. In Silico Experiment Definition: The possibility to use previous knowledge about available approaches to solve the problem is an added value of these portals.
2. Application Design: The application should leverage available tools and hide execution details such as options selection, parameter passing, data format conversion, and so forth.
3. Application Execution: Application should be performed under different resource availability conditions.
4. Result Collection, Visualisation, and Annotation:
Results need to be visualised and analysed and possibly annotated, to provide the so called “provenance” data, that is, information about origin and history of the data.
A common approach for the distributed execution of applications is the service-oriented architecture (SOA) where autonomous software programs (Web services) can be searched, composed and executed by using standard protocols provided by the World Wide Web Consortium (Erl, 2001). In the following a bioinformatics Web portal based on the service-oriented architecture is presented.
PROTEUS: A BIOINFORMATICS WEB PORTAL
PROTEUS is a bioinformatics Web portal based on the problem-solving environment (PSE) approach, useful to define, describe and execute distributed applications (Gallopoulos, Houstis, & Rice, 1994). The top layer of such a PSE represents a scientific Web portal, that is, the user interface through which to access and coordinate different basic bioinformatics components and data banks.
A PSE provides software tools and assistance to the scientist for running applications in a user-friendly environment. This helps in running applications, testing ideas and providing high-performance computing resources. Users are thus relieved from computational details and they may concentrate on the application. PSE is typically aimed at a particular application domain, and cannot be generally used in different application domains without redesigning and reimplementing most or all the environment. The combination of the PSE approach with Web portal techniques and ontology modeling, led to the design of PROTEUS (Cannataro, Comito, Congiusta, & Veltri, 2004), a software architecture allowing building and executing bioinformatics applications on the grid. To help scientists in bioinformatics research, PROTEUS models with ontologies bioinformatics processes and resources such as: (1) biological databases; (2) bioinformatics tools and software; and (3) bioinformatics processes. Ontologies (Gruber, 1993) form a bioinformatics knowledge base, representing knowledge about (biomedical) resources and processes.
In summary, PROTEUS uses ontologies for modeling bioinformatics processes and grid resources, and workflow techniques for designing and scheduling bioinformatics applications, with the aim to assist users in:
• formulating problems, allowing comparison of different available applications to solve a given problem, or to define a new application as composition of available software components;
• running an application on the grid, using the resources available in a given moment thus leveraging the grid scheduling and load balancing services;
• viewing and analysing results, by using high-level graphic libraries, and accessing the past history of executions, that is, the past results that form a knowledge base.
Architecture
PROTEUS combines existing open source bioinformatics software and public-available biological databases by: (1) adding metadata to software; (2) modeling applications through ontology and workflows, and (3) offering prepackaged grid-aware bioinformatics applications. In particular PROTEUS comprises:
• A Web-based Graphical User Interface that allows the search and composition of bioinformatics Web/grid services. The semantic of tools and data sources is modeled through ontologies whereas workflows are used to describe complex applications as composition of simpler services.
• A communication and cooperation layer, that is, Internet and more recent computational grids, due to their security, distribution, service orientation, and computational power.
• A collection of Web/grid services, that are used to implement new or to wrap existing bioinformatics tools.
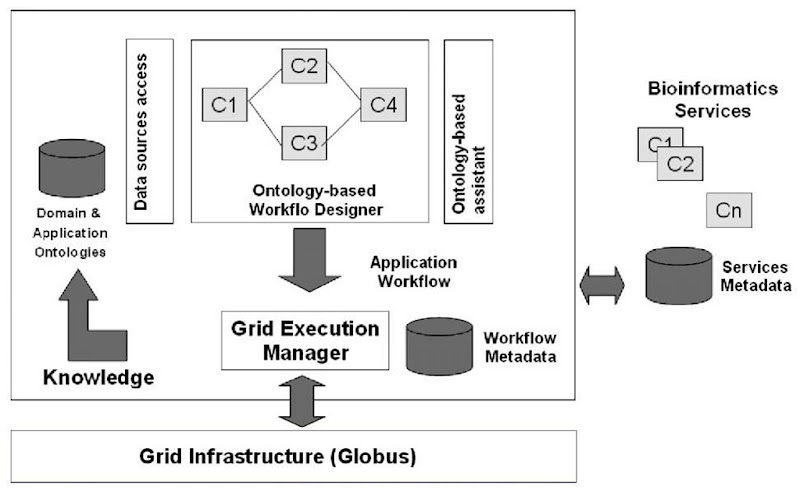
Main components of PROTEUS architecture (see Figure 1) are:
• Metadata Repository: About technical details of software components and data sources.
• Ontologies: We have two kinds of ontology in our system: a domain ontology and an application ontology. The former describes and classifies biological concepts and their use in bioinformatics as well as bioinformatics software tools and biological databases.
Figure 1. PROTEUS architecture
The latter describes and classifies main bioinformatics applications, represented as workflows, and contains information about the application’s results.
• Ontology-Based Workflow Designer: An ontology-based assistant either suggests to the user the available applications for a givenbioinformatics problem/task, or guides the application design through a concept-based search of basic components (software and databases) into the knowledge base. Selected software components are composed as workflows through graphic facilities.
• Workflow-Based Grid Execution Manager. Graphic representations of applications are translated into grid execution scripts for grid submission, execution, and management.
The PROTEUS Web Interface: Ontology-Based Workflow Designer
The main goals of a bioinformatics portal is to help the user in defining and designing the application and to hide the details of the execution environment. Here we focus on the former aspect, since the composition of an application is logically more close to the user than application scheduling. A main role in simplifying application design in PROTEUS is its ontology-based workflow designer, that is, the upper component shown in Figure 1. Such component is made available as a Java Applet through the bioinformatics Web portal. In other words, using the ontology-based workflow designer, the user has a semantic view of all tools managed by the system and can, interactively, design bioinformatics workflows using a Java-enabled Web browser.
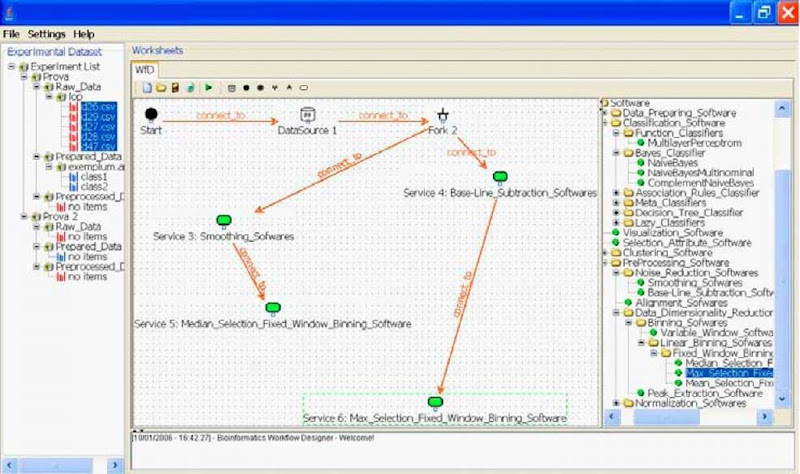
Figure 2 reports the screen shot of the workflow designer user interface running on a grid node. In particular, the right pane contains the ontology-based assistant that shows the PROTEUS ontologies and allows the choosing and selection of available bioinformatics tools. The left pane shows available biological datasets. The middle pane contains the proper workflow editor that allows the designing of the bioinformatics application by combining bioinformatics tools and data sources through workflow constructs.
Figure 2 shows a fragment of a bioinformatics workflow where a data source (a mass spectrometry dataset) is pre-processed in parallel by using two different approaches: (1) smoothing and median-selection binning; and (2) baseline subtraction and max-selection binning. In the rest of the workflow (not shown in Figure 2), the two resulting prepro-cessed datasets are further analysed by using a data mining tool selected on the right pane, to evaluate the impact of the different preprocessing techniques on quality of data mining or on execution performance. Once the workflow has been designed, it is translated into an execution plan containing information on grid nodes hosting services or data.
FUTURE TRENDS
Currently, many problems in bioinformatics are related to the huge volume of data that is produced by biological experiments and that has to be managed and analyzed by bioinformatics tools. In mass spectrometry, for instance, tens of instruments may be able to generate thousands of experimental data results each day. Such data need to be used as input for querying database containing theoretical experimental results, and to identify proteins. Currently such results are only partially stored, for instance, focusing on particular kinds of experiments (see SDBS, 2004). Providing data sources containing also experimental data may become mandatory for comparing results among distributed laboratories. In such a direction, grid infrastructure can be used for sharing data among biomedical laboratories.
Figure 2. The ontology-based workflow designer
CONCLUSION
In such a chapter, we surveyed main bioinformatics platforms and current bioinformatics Web portals and described the requirements for distributed bioinformatics applications. We then discussed the problem of defining a bioinformatics Web portal for designing and running bioinformatics applications on the grid, that can be thought of as a virtual laboratory joining remote bioinformatics resources. We proposed the PROTEUS environment that combines a Web-based graphical user interface and an ontology dictionary. It allows biological experts to use and compose applications and data sources concentrating on the application logic instead of technical details.
KEY TERMS
Bioinformatics: Bioinformatics involves the use of techniques from applied mathematics, informatics, statistics, and computer science to solve biological problems.
Bioinformatics Web Portal: Is a software platform accessible through a Web portal specifically designed to execute bioinformatics applications.
Biological Databases: A database containing data describing biological elements, for example, proteins, nucleotides, and so on.
Grid: A grid is a computing infrastructure that uses the resources of many separate computers connected by a network (usually the Internet) to solve large-scale computation problems.
“In silico” Experiment: Is “an experiment performed on computer or via computer simulation.” The phrase is coined from the Latin phrases in vivo and in vitro that are commonly used in biology and refer to experiments done in living organisms and outside of living organisms, respectively.
Ontology: In computer science an ontology is a data model that represents a domain and is used to reason about the objects in that domain and the relations between them.
Problem Solving Environment: Is a software platform enabling the design and execution of applications in a specific domain.
Workflow: A workflow is the operational aspect of a work procedure: how tasks are structured, who performs them, what their relative order is, how they are synchronised, how information flows to support the tasks and how tasks are being tracked.