Abstract
Bone image segmentation is an integral component of orthopedic X-ray image analysis that aims at extracting the bone structure from the muscles and tissues. Automatic segmentation of the bone part in a digital X-ray image is a challenging problem because of its low contrast with the surrounding flesh, which itself needs to be discriminated against the background. The presence of noise and spurious edges further complicates the segmentation. In this paper, we propose an efficient entropy-based segmentation technique that integrates several simple steps, which are fully automated. Experiments on several X-ray images reveal encouraging results as evident from a segmentation entropy quantitative assessment (SEQA) metric [Hao, et al. 2009].
Keywords: Entropy, Digital X-ray, LOCO, Medical imaging, Segmentation.
Introduction
Segmentation of an anatomical object from an X-ray, MRI or a CT-scan image is a challenging task as the intensity values of the region-of-interest and the surrounding region are very close, and the contour clarity is often low. Many traditional image segmentation approaches are computationally intensive or need manual intervention [1, 2].
In last two decades, numerous segmentation models and algorithms for analyzing various classes of medical images have been reported. The selection of proper segmentation method is another important issue as this choice widely depends on specific application, image modality and several other factors [1, 2]. In this paper, we briefly discuss some notable segmentation algorithms used in medical imaging and then propose an integrated entropy-based technique suitable for bone segmentation in a digital X-ray image from the surrounding flesh region. The method is simple and fully automated and it uses a new thresholding parameter. The quality of segmentation is observed to outperform those obtained by several earlier methods [3, 13] as indicated by the segmentation entropy quantitative assessment (SEQA) [16].
Medical Image Segmentation: Prior Art
Image segmentation is defined as its partitioning into different parts, which are homogenous in nature with respect to some characteristics. The region-of-interest (ROI) may be a complete object or a part of it [2, 5]. Medical image segmentation algorithms can be classified into categories like those based on analysis of edges, regions, pixel classification, graphs, or on deformable model, fuzzy logic, artificial neural network, statistical or morphological approaches. The segmentation algorithms based on region analysis or pixel classification, make use of the homogeneity in the image, whereas the edge detection and graph-based algorithms perform segmentation by detecting any abrupt change in the image within a small neighborhood [5]. The Canny edge detector uses two levels of thresholding. Scepanovic et al. adopted Canny’s method for probabilistic bone edge detection [6].
The deformable model based approaches allow elastic decomposition of the objects by incorporating concepts of continuum mechanics. The active contour or snake model was proposed by Kass et al. [7]. In deformable models like snake, the user provides an initial guess of contour of ROI, which is then moved by the image-driven forces to the boundary of the desired object. However, the snake method with gradient vector flow (GVF) has slow convergence on irregular objects with high concavities like X-ray images of deformed bone structure.
The active shape model and active appearance model are also used in medical image segmentation. Unfortunately, both the methods require large training samples to build a point distribution in the high-dimensional eigen space. The statistical models like probability distribution map and Markov random field model have also been used widely for medical image segmentation. Liang et al. have used statistical and morphology based technique for fracture identification in an X-ray image.
Several semi-automatic image segmentation methods incorporate user interaction and allow the user to mark the ROI or background objects roughly on the image. Ning et al. proposed an interactive image segmentation algorithm based on maximal similarity based region merging [12].
Thresholding-based segmentation approaches are widely used in medical image analysis [8, 9]. Sen et al. proposed a method based on gradient histogram and applied it on a chest X-ray image among others [8].Yan et al. used an entropy-based thresholding approach for segmentation of bacteria images from a low intensity background [13]; however, this method requires a tuning parameter, which is chosen based on experience.
Proposed Approach for X-ray Image Segmentation
In an X-ray image, the bone parts appear along with the surrounding tissues or muscles (i.e. flesh) image. In many regions of an X-ray image, the intensity range of pixels belonging to the bone region and that of its surrounding flesh region may overlap, which makes the segmentation inaccurate. The bone region of an X-ray image is to be extracted out of two distinct background components, the image background and the overlapping flesh region. Thus, bone segmentation from the flesh region using pixel based thresholding or edge based approaches often do not produce accurate results.
In this paper, we have proposed a new entropy-based approach, which instead of direct entropy thresholding [13], first produces an intermediate image from the entropy matrix and then uses intensity-thresholding for segmenting the bone region in an X-ray image. Finally, for contour correction, multilevel LOCO (Linear combination of Open-Close and Close-Open) filtering methods [11] are used.
Integrated Segmentation Algorithm
In the proposed algorithm, the transition regions in an input image are identified using entropy analysis and a gray scale entropy image is generated. This entropy image is used to compute the threshold value required for segmentation.
A high entropy value represents the presence of maximum information, i.e., higher gray value pixels in the image. An image with a very low entropy value represents noise with no meaningful information. The overlapping flesh and bone regions in an X-ray image cause the presence of a large number of pixels with similar intensity values. Computation of local entropy values of each pixel in a window helps to group the pixels with similar intensity values with a marked transition region. As a bone region has higher intensity values than those in the flesh region in an X-ray image, the transition boundaries are accentuated in the entropy image. The steps of the proposed algorithm are summarized below, details of which are elaborated subsequently.
Step 1: Convert an input X-ray image to a gray scale image.
Step 2: Perform entropy computation of each image pixel around (9×9) neighbors. For pixels at the border of the image, symmetric padding is used where the padded pixels are obtained by mirror reflection of the border pixels of the image.
Step 3: Generate an entropy image by converting the entropy matrix to a gray scale image, where the maximum entropy value is mapped to 255 and the minimum entropy value is mapped to 0. Other entropy values are linearly scaled between 0 and 255.
Step 4: Apply median filter on the entropy image.
Step 5: Compute the intensity threshold value for binarization of the entropy image. Step 6: Produce the segmented bone image.
Step 7: Apply multilevel LOCO filter to enhance the contour of the segment.
Entropy Computation and Constructing Entropy Image
The entropy value represents the measure of randomness associated with pixel values belonging to each gray level.
Following Shannon’s definition of entropy and Pun’s representation of image entropy, for an image with G gray levels, and probability of k-th gray level Pk, the image entropy can be represented as
where Pk = nk/MxN , nk is the number of pixels with grayscale k and MxN is the size of the image [5].
Local entropy represents the discrepancy between two probability distributions on the same event space [13]. It is related to the variance of grayscales in the neighborhood of a pixel. Local entropy divides the image into separate regions and then analyzes each region as a separate information source. If a small neighborhood window![]() of size
of size ![]() is defined within the image, then entropy of
is defined within the image, then entropy of![]() can be represented as:
can be represented as:
where, Pj denotes the probability of gray scale j in the neighborhood![]() is the number of pixels with gray scale j in
is the number of pixels with gray scale j in![]() the local entropy of
the local entropy of![]()
Yan et al. had applied local entropy based thresholding for transition region extraction [13]. It was shown that the local entropy is larger for heterogeneous regions and smaller for homogeneous neighborhoods. So, a transition region will have larger local entropy values than those in the non-transition regions of the image. This boundary enhancement aids thresholding for segmentation of the transition region from the image. Kang et al. used local entropy values to identify transition regions on region extraction from coronary angiograms [14].
In the proposed algorithm, we calculate the local entropy values by taking a (9×9) neighborhood window for each pixel of the X-ray image. The pixels at the transition region enhance their entropy values compared to those of other pixels.
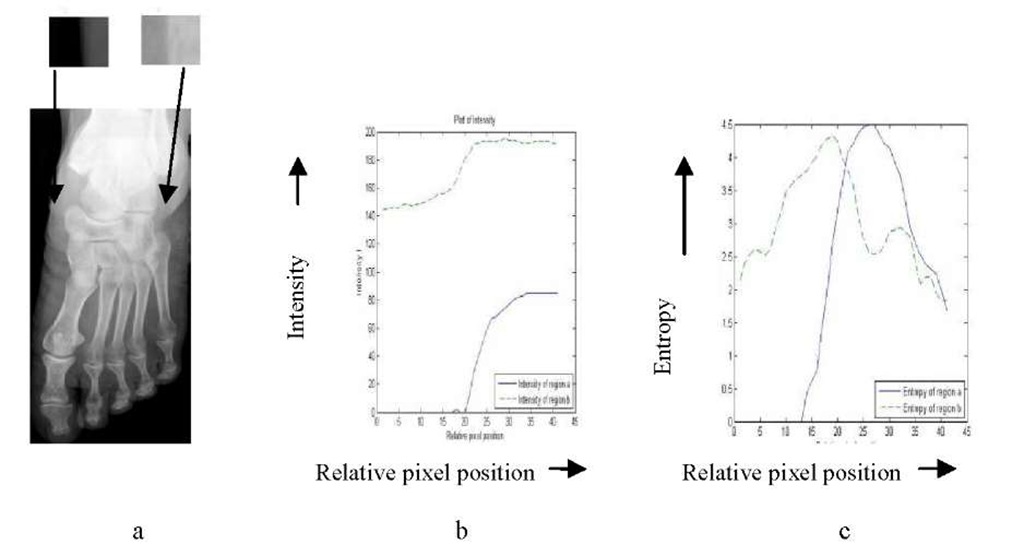
In an X-ray image of foot (Fig. 1a), the intensity variation of the transition region s1 ranges between 0 to 80, whereas, for the transition region s2, it varies from 150 to 190 (Fig. 1b). In Fig. 1c we show the variation in entropy values for the transition regions s1 and s2. It is observed that the entropy values at transition region show a high peak irrespective of the difference in the intensities at the transition regions. We have used this entropy behavior to construct an intermediate gray scale image (called entropy image) in which the bone boundaries appear at high entropy regions (i.e., transition regions). Next, an intensity thresholding approach is applied to segment the bone.
Fig. 1. a: Gray scale image of foot X-ray with transition regions s1 and s2, b: Intensity variation in s1 and s2, c: Entropy variation in s1 and s2
Morphological Filter: Multilevel LOCO
Since X-ray image processing is often corrupted by noise and spurious edges, we use a morphology-based special filtering technique to improve the contour definition of the segmented bone parts. Morphological opening is defined as erosion followed by dilation and closing is dilation followed by erosion using some structuring element [5].
A LOCO filter (Linear combination of Open-Close (OC) and Close-Open (CO)) is designed by averaging the cascaded opening and closing operations within a generalized filter structure [11]. The LOCO filtering on an image I with a structuring element N can be represented as:
The segmented bone structure obtained using the proposed algorithm may suffer from having thin or disconnected boundaries. A LOCO filter with multiple structuring elements is applied to enhance the bone boundary and to improve the visual quality of the segmented bone image. We use three different (3x 3, 5×5 and 7×7) diamond-shaped structuring elements [5] and generate enhanced contour image using each of them. Finally, the output image is produced by taking the average of these pixel values.
Thresholding
The OTSU method is a well known thresholding algorithm, which selects the threshold by minimizing within class the variance between two groups of pixels separated by thresholding operator [3]. However, this method may not work well if the two classes are very unequal and if the image has variable illumination. This is a very common problem in an X-ray image. So we have used global thresholding approach based on mean intensity value to determine the threshold for bone image segmentation. In this approach, the threshold value is determined by computing the maximum of the mean intensity value for each column of the image matrix and the mean intensity value for each row; this value can then be used for binarization of the gray scale X-ray image.
Experimental Results
We have run the proposed algorithm on 40 digital X-ray images of different portions of human body, collected from the literature and other sources. For lack of space, the output generated only for a few of them are shown in this section (Figs. 2, 3).
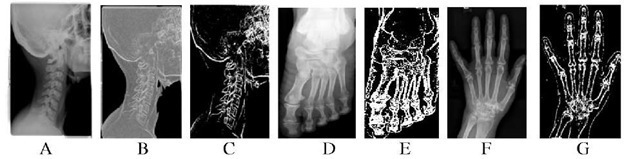
Fig. 2. A: Input X-ray image of throat, B: Gray image of A after entropy analysis, C: Segmented bone image by the proposed algorithm, D: Input X-ray image of foot, E: Segmented bone image, F: Input X-ray image of broken wrist, G: Segmented bone image of F using the proposed method
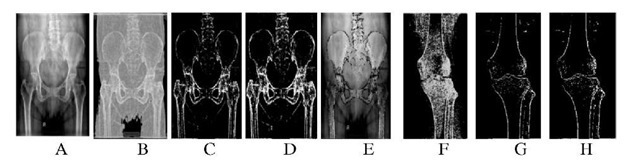
Fig. 3. A: Input X-ray image of pelvic bone, B: Gray image of A after entropy analysis, C: Segmented bone image after thresholding, D: Segmented bone image with bone boundary enhanced by applying multilevel LOCO, E: Segmented image superimposed on original image, F: Segmented knee image for a = 0.7, G: Segmented knee image for a = 0.8, H: Segmented knee image using the proposed algorithm
The proposed segmentation algorithm is compared with another entropy-based thresholding approach proposed by Yan et al. [13]. The idea of local entropy thresholding approach was applied on several bacteria images for segmenting them from the background. The thresholding value is computed as a times maximum entropy, where a is a coefficient between 0 and 1.
where![]() is the maximum entropy value for region
is the maximum entropy value for region![]() .In this approach a is empirically chosen between 0.6 to 1.0, which requires manual intervention. In the proposed X-ray segmentation approach, the entropy matrix of the input image is converted into a gray scale entropy image and thresholding is performed after binarization. Hence, this approach is fully automated and does not need any training samples.
.In this approach a is empirically chosen between 0.6 to 1.0, which requires manual intervention. In the proposed X-ray segmentation approach, the entropy matrix of the input image is converted into a gray scale entropy image and thresholding is performed after binarization. Hence, this approach is fully automated and does not need any training samples.
Figs. 3F, 3G, 3H reveal that the segmentation of the bone boundary of knee image is more accurate for a = 0.8, whereas for a pelvic bone image a = 0.7 gives better results when the earlier algorithm [13] is applied. The proposed segmentation method yields an improved result automatically (Fig. 3H).
Quantitative Performance Evaluation
We have evaluated the performance of the proposed algorithm using a recent segmentation entropy quantitative assessment (SEQA) method, which is based on segmentation entropy to compute an evaluation metric [16]. Another evaluation method called ultimate measurement accuracy (UMA) [17], uses the area of segmented object as a powerful evaluation metric. However, in the case of a bone image, this is difficult to compute because of fragmentation of contours. Thus, we have used the SEQA method for our purpose. In this approach, if the segmentation process S divides an image I into n subregions and E(ri) represents the entropy of ri th region, the segmentation entropy E(S) is defined as: ![tmp1411-1629_thumb[2][2] tmp1411-1629_thumb[2][2]](http://what-when-how.com/wp-content/uploads/2011/07/tmp14111629_thumb22_thumb.jpg)
where E(I) represents the total entropy of the original image.The segmentation entropy E(S) measures how much the image entropy changes during segmentation. Better segmented regions will be more homogeneous in nature and thus they will have lower region entropy. If the same image is segmented using two different segmentation algorithms A and B, and if E(A) is smaller than E(B), then according to Eqn (5), the performance of A is better than that of B [16].We have compared the segmentation entropy E of the proposed method with those of OTSU thresholding [3] and entropy thresholding [13]. These values for different digital X-ray images as listed in Table 1 support the effectiveness of the algorithm.
Table 1. Computation of segmentation entropy for some digital X-ray images
|
Image file |
E(S) for OTSU |
E(S) for entropy thresholding; a = 0.7 |
E(S) for proposed method |
|
Finger-Left.jpg (Fig. 2F) |
3.52 |
2.63 |
2.05 |
|
Foot.jpg (Fig. 2D) |
2.09 |
0.80 |
0.83 |
|
Leg.jpg (Fig. 3F) |
2.48 |
1.92 |
1.48 |
|
Pelvic Bone.jpg (Fig. 3A) |
1.77 |
0.82 |
0.77 |
|
Throat.jpg (Fig. 2A) |
1.11 |
0.30 |
0.28 |
Discussion and Conclusion
We have developed an entropy-based X-ray image segmentation technique for bone part extraction. This is based on automatic thresholding of an entropy image defined by the entropy values in a neighborhood window of the given image. It has been observed that in many cases the bone boundary becomes fragmented after thresholding. To overcome this problem, we have applied a multilevel-averaging LOCO filter to enhance the bone contour of the segmented image. The method is fully automated and has superior performance over two earlier methods as evident by the SEQA metric. This algorithm will be useful for fracture detection in an X-ray image where, separation of bone from the surrounding flesh is needed for further analysis.
![tmp1411-1606_thumb[2][2] tmp1411-1606_thumb[2][2]](http://what-when-how.com/wp-content/uploads/2011/07/tmp14111606_thumb22_thumb.png)
![tmp1411-1613_thumb[2][2] tmp1411-1613_thumb[2][2]](http://what-when-how.com/wp-content/uploads/2011/07/tmp14111613_thumb22_thumb.jpg)