The nucleotide-binding motif is a structural and functional domain that is frequently observed in protein structures that bind nucleotides. It has a characteristic sequence that can usually be observed in the primary structure. There are two classes of the motif; the classical dinucleotide-binding fold and the classical mononucleotide-binding fold. The former binds dinucleotides such as NAD and FAD, and the latter binds mononucleotides such as ATP (and is also called the ATP-binding motif).
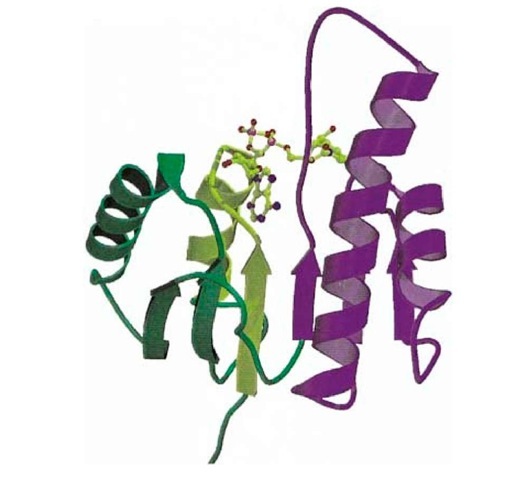
The dinucleotide motif was first identified by Rao and Rossmann, who identified two b-a-b-a-b units (b is a b-strand and a is an a-helix) forming a six-stranded parallel b-sheet surrounded by four a-helices in the structures of lactate dehydrogenase (LDH) and malate dehydrogenase (Fig. 1) (1). Both bind NAD; each half of the dinucleotide interacts with one b-a-b-a-b unit. A single b-a-b-a-b unit is called a Rossmann fold. The adenine moiety of NAD (or FAD) binds to the first of the two b-a-b-a-b- units, and the nicotinamide (or flavin) binds to the second. The first three elements of secondary structure in the adenine-binding motif, b1-aA-b2, are involved in the predominant interactions with adenine and have a common structure and related sequences (1). This "fingerprint" sequence consists of conserved small hydrophobic residues in b1, followed by a glycine-rich region (-Gly-X-Gly-X-X-Gly, where X is any residue), conserved small hydrophobic residues in helix aA, and an Asp or Glu at the end of helix aA. The first two glycine residues are in a loop connecting b1 and aA; the third glycine is in helix aA. The glycine residues adopt conformations forbidden to other residues and allow close packing of the b-strands and helix and a close approach between the adenine pyrophosphate and the ^-terminal end of the aA-helix. The phosphate group is thought to interact favorably with the partial positive charge at the ^-terminal end of the aA-helix dipole. The conserved Asp/Glu residue interacts with the ribose hydroxyl. This signature sequence of ~30 residues was derived from the tertiary structures of known NAD-binding proteins, but it can also be used to predict NAD- or FAD-binding proteins from their primary structure.
Figure 1. Schematic representation of the backbone structure of the nucleotide-binding domain of malate dehydrogenase (1). b-Strands are shown as arrows and a-helices are shown as coils. The bound dinucleotide (NAD) is shown as a ball-and-stick model. The two b-a-b-a-b- units that interact with the each half of the mononucleotide are shown in green and purple. The b1-aA-b2 region that interacts with the adenine portion of NAD is shown in yellow. This figure is generated using Molscript (2) and Raster3D (3, 4).
The fingerprint sequence is slightly different for binding the dinucleotide NADP, where the 2′ ribose hydroxyl of NAD is phosphorylated. In this case, the third glycine (in the aA-helix) is replaced by Ala so that the close approach of aA and the b-strands is prevented. This provides additional space for binding the 2′-phosphate. Furthermore, the conserved Asp/Glu of the NAD/FAD sequence is replaced by Arg to interact with the 2′-phosphate.
The classical mononucleotide or ATP-binding motif, typified by adenylate kinase, is also an a/b protein fold but has a five-stranded parallel b-sheet with a very different connectivity from the dinucleotide fold. Once again, the bound nucleotide binds to a b1-aA-b2 region that has a characteristic fingerprint sequence: Gly-X-X-Gly-X-Gly-Lys. In this case, however, the glycine-rich sequence corresponds only to the loop between b1 and aA, so that all three glycine residues form part of the loop (in contrast to the dinucleotide sequence, where the third glycine is part of helix aA). The loop forms a large pocket that accommodates the phosphate of the mononucleotide (it is also called the P-loop), and the glycine amides and the side chain of the conserved lysine residue interact with the phosphate.