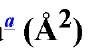The surfaces of folded macromolecules, especially proteins, and the internal packing of their atoms have generally been analyzed using the procedure of Lee and Richards (1). The cross section of part of the surface of a native protein is depicted in Figure 1, which demonstrates a number of different surfaces and volumes (see Protein Structure). The van der Waals surface is defined by the spherical atoms that comprise the structure, but it is not very relevant to a folded macromolecule like a protein, where internal atoms and cavities are normally inaccessible to the solvent.
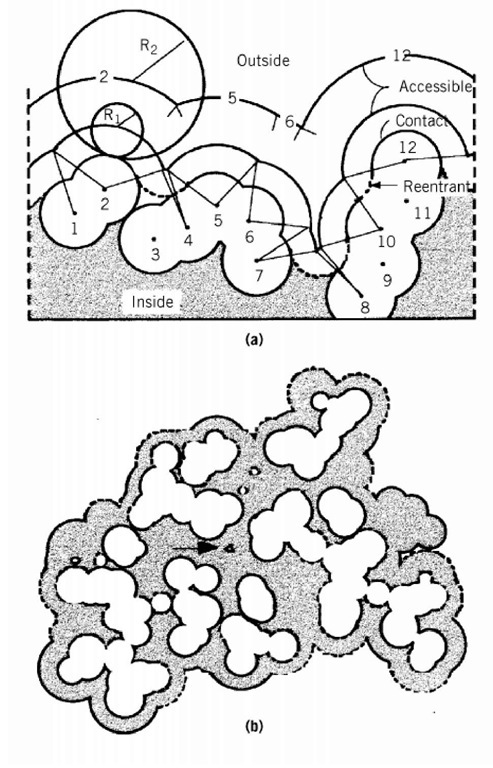
Figure 1. Schematic representation of possible molecular surface definitions. (a) A two-dimensional section through part of the van der Waals envelope of a hypothetical protein including 12 atoms with the centers numbered 1-12. Rx and R2 show the radius of the probes. (b) Superposition of sections through the van der Waals and accessible surfaces of ribonucrease S. In places, the accessible surface is controlled by atoms above or below the section shown. The solid outline is the surface of carbon and sulfur atoms; the dashed outline nitrogen and oxygen. The arrow indicates a cavity inside the molecule large enough to accommodate a solvent molecule with a radius of 1.4 A.
The most relevant surface is the accessible surface, which is defined by its contact with molecules of the solvent. This surface is defined by rolling a spherical probe of appropriate radius R^ on the outside of the molecule, while maintaining contact with the van der Waals surface. For most purposes, the appropriate probe is a water molecule. The accessible surface is that depicted by the center of the probe as it moves over the surface of the protein. In Figure 1 the probe does not contact atoms 3, 9, or 11, and they have no accessible surface area. Such atoms are considered to be interior atoms, not part of the surface of the molecule. Those parts of the van der Waals surface in contact with the surface of the probe are designated the contact surface; they comprise a series of disconnected patches. When the probe is simultaneously in contact with more than one protein atom, its interior surface defines the reentrant surface. The contact surface and reentrant surface together make a continuous surface, which is defined as the molecular surface.
The accessible surface area depends on the size of the probe. When the radius of the probe increases from R1 to R2, the number of noncontact or interior atoms in Figure 1 increases from three to eight, and the accessible surface is much smoother. Thus, the smaller the probe, the larger the number of features that will be revealed. The probe is frequently taken to be a water molecule and approximated as a sphere with a radius of 1.4 A. The accessible surface areas of individual amino acid residues are given in Table 1.
Table 1. Accessible Surface Areas of Amino Acid Residues
|
Ala |
113 |
|
Arg |
241 |
|
Asn |
158 |
|
Asp |
151 |
|
Cys |
140 |
|
Gln |
189 |
|
Glu |
183 |
|
Gly |
85 |
|
His |
194 |
|
Ile |
182 |
|
Leu |
180 |
|
Lys |
211 |
|
Met |
204 |
|
Phe |
218 |
|
Pro |
143 |
|
Ser |
122 |
|
Thr |
146 |
|
Trp |
259 |
|
Tyr |
229 |
|
Val |
160 |
a Values estimated for a Gly-X-Gly tripeptide in an extended conformation (3)
The structures of protein determined by X-ray crystallography indicate that the total accessible surface area![]() of a protein is approximately proportional to the two-third power of its molecular weight. The
of a protein is approximately proportional to the two-third power of its molecular weight. The![]() of a typical small monomeric protein is usually related to its molecular weight
of a typical small monomeric protein is usually related to its molecular weight![]() by the approximate relationship (2)
by the approximate relationship (2)
For oligomeric proteins
These equations are usually accurate to within ±4% on average for monomers and 5% for oligomers. Equations 1 and 2 imply that an oligomeric protein has a larger accessible surface area (by 7 to 13%) than a monomeric protein of the same molecular weight in the molecular weight range up to 35,000.
For an unfolded polypeptide chain, the total accessible surface area At in an extended conformation is directly proportional to its molecular weight, within ±3%
Thus, for those proteins whose accessible surface area is given by Equation 1, the potential surface area buried by the protein’s folding Ab will be approximately given by
This indicates that 55 to 75% of the surface area of the unfolded polypeptide chains is buried in the folding process.
The methodology of Lee and Richards (1) is also applicable to the calculation of the accessible surface area of nucleic acids (4). Two-thirds of the water-accessible surface area become buried on double-helix formation of DNA and RNA. When a probe corresponding to a single water molecule is used, the total accessible surface area is similar for A-DNA and B-DNA, although marked differences appear in the major and minor groove exposures. For the larger probes, there exist considerable differences in accessible surface area between the two conformations.