Abstract
By bringing people the right information in the right format at the right time and place, state of the art clinical information systems with imbedded clinical knowledge can help people make the right clinical decisions. This topic provides an overview of the efforts to develop systems capable of delivering such information at the point of care. The first section focuses on “library-type” applications that enable a clinician to look-up information in an electronic document. The second section describes a myriad of “realtime clinical decision support systems.” These systems generally deliver clinical guidance at the point of care within the clinical information system (CIS). The third section describes several “hybrid” systems, which combine aspects of real-time clinical decision support systems with library-type information. Finally, section four provides a brief look at various attempts to bring clinical knowledge, in the form of computable guidelines, to the point of care.
To be effective, (clinical decision support) tools need to be grounded in the patient’s record, must use standard medical vocabularies, should have clear semantics, must facilitate knowledge maintenance and sharing, and need to be sufficiently expressive to explicitly capture the design rational (process and outcome intentions) of the guideline s author, while leaving flexibility at application time to the attending physician and their own preferred methods. (Shahar, 2001)
Introduction
By bringing people the right information in the right format at the right time and place, informatics helps people make the right clinical decisions. Cumulatively, these better decisions improve health outcomes, such as quality, safety, and the cost-effectiveness of care. This improvement has been a mantra of informatics at least since the landmark article by Matheson and Cooper in 1982. This topic provides an overview of the efforts over the years to develop systems capable of delivering such information at the point of care. Such an overview should help illustrate both the opportunities and challenges that lie ahead as we struggle to develop the next generation of realtime clinical decision support systems for use at the point of care.
vision to achieve
The ultimate goal is to provide patient-specific, evidence-based, clinical diagnostic and therapeutic guidance to clinicians at the point of care; this guidance should be available within the clinical information system (CIS) that defines their current workflow. In addition, we must have the tools necessary to enable clinicians, without specialized programming knowledge, to enter, review, and maintain all the clinical knowledge required to generate this advice. Finally, we must have the ability to rapidly change the clinical knowledge, test it, and make it available to clinicians without having to wait for a regular CIS updating schedule.
Questions That Must Be Answered prior to creating such systems
What information or knowledge is required to help the clinician make the right decision to achieve the desired health outcome?
Who will be the information’s recipient (e.g., physician, nurse, pharmacist, or even a specific individual such as the patient’s primary care physician)?
When in the patient care process is the intervention applied, for example, before, during (which can be broken down into sub-activities such as order entry or progress note creation) or after the patient encounter?
How is the intervention triggered and delivered? For example, does the system or the clinician initiate it? How much patient-specific data (if any) is needed to trigger system-initiated interventions? How much is the intervention output customized to the clinical workflow stage, the clinician, and or the patient? For system-initiated interventions, how can the threshold be set to minimize nuisance alerts? How intrusive should the information be (Krall, 2001)?
Will the clinicians find the information useful (Krall, 2002a, 2002b)? What can we do to minimize the number of false positive alerts?
Where will the clinician be when receiving the intervention, for example, with the patient at the bedside or in the office? What should happen if the information becomes available at some future point when the clinician is no longer with the patient to whom the information pertains?
Which medium will be used to convey the message, for example, e-mail inbox, wireless and/or handheld device, pager, CIS/CPOE screen, or printed pre-visit encounter sheet?
Is there a demonstrable return on investment (ROI) that is due exclusively to the clinical decision support intervention or feature?
background where Do we stand?
Numerous attempts have been made to bring various forms of clinical information to the clinician at the point of care. One way to solve this problem is to develop computer applications that stand alone, although often network accessible, and are available to the clinician upon his/her request. Another way is to incorporate the clinical knowledge directly into the clinical information system used by clinicians while giving care. Once it is there, the CIS system can automatically prompt the clinician or the clinician can request help. Recently, a third model for system development has been proposed that enables the clinician to request help from an outside source. Often several patient-specific data items are included in the request. Using such a system, a clinician is still completely in-charge of making the request for information and the information can be automatically configured based on a subset of patient information (Cimino, 1996). In addition, it is now possible to embed medication or procedure codes along with instructions that tell the CIS how to handle this information that allows a clinician to place an order directly into the patient’s electronic medical record from the externally available information resource (Tang & Young, 2000).
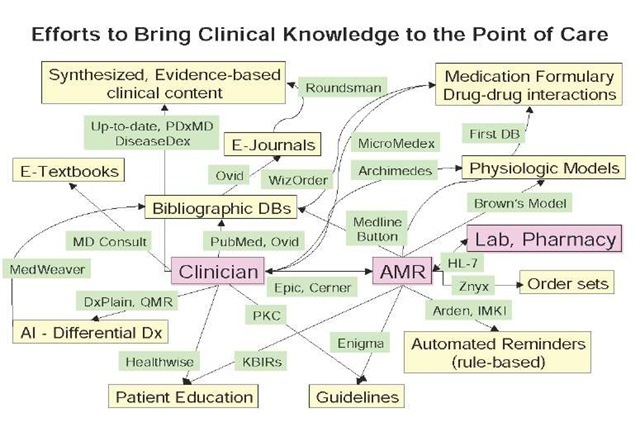
The following diagram (Figure 1) is an attempt to illustrate both the key types of information or knowledge that investigators have focused on along with their mode of interaction [i.e., directly to the clinician or through the clinical information system (CIS)]. The boxes (yellow) represent the type of knowledge and the labels on the links (green) represent some of the key projects or concepts vendors have focused on this particular form of clinical decision support. The aim of this figure is to highlight the myriad attempts that have been made to develop clinical knowledge management applications and to help everyone understand how different clinical knowledge resources and applications are both related in terms of what they are trying to accomplish and different in the resources they utilize. Although this diagram is fairly complex, it is only a small, imperfect and incomplete representation of the entire clinical decision support landscape.
The following sections briefly review each of the projects or concepts. The first section focuses on “library-type” applications that enable a clinician to look up information in an electronic document. The second section describes a myriad of “real-time clinical decision support systems”. These systems generally deliver clinical guidance to clinicians at the point of care within the CIS. The third section describes several “hybrid” systems, which combine aspects of knowledge-based clinical decision support systems with library-type information. Finally, section four looks at various attempts to bring clinical knowledge in the form of computable guidelines to the point of care.
Figure 1. An overview of the numerous efforts researchers have made to bring pertinent clinical knowledge to clinicians at the point of care
Bibliographic Databases (DBs)
Biomedical bibliographic databases contain on the order of millions of records, each representing a unique scientific journal article that has been published. Each record typically contains the title of the article, the authors, their affiliation(s), and the abstract of the article. In addition, several metadata tags may have been applied by human indexers to improve the likelihood that each document will be retrieved when and only when it is relevant to the user’s query.
For the last 30 years, the National Library of Medicine has maintained the MedLine database, the most common bibliographic database used in clinical medicine. During that period, various attempts have been made to develop easy to use and reliable interfaces to this vast resource including Grateful Med (Cahan, 1989) and COACH (Kingsland, 1993). Currently, PubMed is the most widely used of all these interfaces. PubMed relies on a sophisticated free-text query processor to map freetext user queries to MeSH terms, when appropriate, and returns a highly relevant set of documents.
In addition to PubMed, several commercial vendors have created proprietary interfaces to the MedLine database in an attempt to improve either the recall or precision of the user’s queries. For example, Ovid has developed an interesting MeSH mapper and query expander that has gathered outstanding reviews from highly trained librarians. Knowledge Finder has developed a fuzzy mapping algorithm that has also generated some good reviews. Unfortunately, none of these systems consistently enables clinicians to retrieve more than half of all the relevant articles on any particular topic (Hersh, 1998). In addition to these variations on a search interface, several projects have used automated differential diagnosis generators, such as DxPlain, as an interface to the bibliographic DBs. Finally, the Science Citation Index uses the reference list at the end of every scientific article published to generate linked lists of references. Such a scheme can also identify particularly noteworthy articles since these articles are referenced many more times. Interestingly, the Google search engine uses this same concept to generate its index of relevant web sites. It does this by keeping track of the number and quality of referring web sites rather than references at the end of the article (Brin, 1998).
other clinical Reference Materials
In addition to access to the bibliographic literature, clinicians could use other clinical reference information. For example, various systems exist to provide clinicians with access to the complete text and figures contained in textbooks (e.g., MD Consult) or journals (e.g., Ovid) in electronic form. In addition, several commercial companies have begun developing synthesized, evidence-based clinical summaries of the diagnosis and treatment of common clinical conditions [e.g., Up-to-Date, CliniAnswers, DiseaseDex, PDxMD.
REAL-TIME clinical decision support systems
Real-time clinical decision support systems (RCDSS) are fundamentally different from “library” applications in that they interact with the clinician through the CIS. Rather than relying on the user to interpret the text and then make a decision, RCDSS combine the patient’s clinical data with the clinical knowledge to help the clinician reach a decision. At least six distinct methods can provide this type of RCDSS:
1. Medication formularies and drug-drug interaction checkers provide routine checks of all orders entered into the EMR. They can recommend alternative medications based on the formulary and check for potential drug-drug interactions automatically following entry of a medication order assuming they have access to the patient’s list of current medications. FirstDataBank, Micromedex, and Multum are examples of commercial providers of drug-drug interaction checking databases.
2. Automated rule-based reminders are used most commonly to remind clinicians to perform routine health maintenance procedures. The Arden syntax is currently the only “standard” means of encoding these rule-based systems (Arden, 2003). The Institute for Medical Knowledge Interchange (IMKI), another recent entry into this field (IMKI, 2003), recently scaled back their operations while they seek additional funding and support.
3. Condition-specific order sets and charting templates help clinicians remember and facilitate the entry of all related orders or answers to questions asked of a patient with a particular condition. Zynx, a recently formed company, is developing these ready-to-use, condition-specific order sets. In addition, most of the commercially available CISs either offer ready-to-use order sets or provide tools to help clinicians construct these order sets (Franklin, Sittig, Schmiz, Spurr, Thomas, O’Connell, & Teich, 1998).
4. Integrated clinical guidelines allow a clinician to compare a specific patient’s clinical data against a guideline and automatically receive a recommendation. The most advanced example of this type of application is Enigma’s PREDICT application. In an early attempt to develop this sort of functionality, Epic Systems created their “Active Guidelines” application. A major distinguishing feature between the Active Guidelines (AG) effort and that of Enigma is that the AG user is still required to search for, read, interpret, and then decide which portion of the large, text-based guideline document applies to a specific patient and then select the appropriate recommendation. Enigma’s PREDICT automatically does all of this work and simply presents the clinician with the appropriate suggestions.
5. Complex, physiologic models incorporate our best current understanding of human physiology along with evidence-based results of randomized clinical trials to generate patient- or population-specific predictions of future events. Archimedes, a complex model of diabetes and the entire healthcare system, is currently configured to allow specially trained clinicians to access this resource (Schlessinger, 2002). Jonathan Brown, from Kaiser Permanente’s Center for Health Research in Portland, has recently begun working on a method to automatically link therapeutic suggestions generated by his Global Diabete s Model to a specific patient’s clinical data from an EMR or clinical data warehouse (Brown, 2000).
6. Artificial Intelligence (AI) systems have been developed to help clinicians complete their differential diagnosis list. To date these systems have not had wide-spread acceptance by clinicians, even though they have been shown to be at least as good and often better than un-aided clinicians (Bankow-itz, 1989; Berner, 1994). DxPlain, QMR, and Weed’s Problem Knowledge Couplers (Weed, 1986) are the best examples of these types of systems.
HYBRID applications
In addition to each of these fairly straightforward attempts to link clinical knowledge to the point of patient care, several companies have attempted to combine various clinical knowledge resources and applications to generate even more useful applications. MedWeaver (Detmer, 1997) was an early attempt to combine DxPlain, an AI-based differential diagnosis (DDx) system with a bibliographic database. The DDx in conjunction with the Unified medical Language system (UMLS) act as an automatic query expander and presents the user with information related to all the known items on the patient’s differential diagnosis.
Geissbuhler and Miller (1998) developed WizOrder, a system to automatically generate potential drug-drug interaction reminders when clinicians order medication through the system. These reminders are based on the co-occurrence of clinical term frequencies1 that are a part of the UMLS’s Metathesaurus database. Rennels, Shortliffe, Stockdale, and Miller (1987) developed Roundsman, a prototype system, to automatically extract evidence-based recommendations from on-line structured journal articles.
Cimino (1996) developed the concept of the “InfoButton” which allows a clinician to automatically generate queries to a variety of clinical information resources to answer a set of generic concept-specific questions such as, “What’s the treatment for X?”, where X can be any clinical condition. Recently, the HL-7 Clinical Decision Support Task Force has begun efforts to develop a standard for such queries.
computable guidelines current state of the Art
Various research and commercial efforts are underway in an attempt to create systems to achieve the vision outlined above. The distinguishing feature of computable guideline systems is that they are more than a simple application of “if-then-else” rules to generate alerts and reminders at the point-of-care. All of the systems described below are attempting to go one step further with multi-step clinical algorithms at the point-of-care. The more complex algorithms attempt to track a patient’s treatments and progress over time, while the alerts and reminders serve only to look at what is best at each point in time. Therefore, I did not include the efforts of the Institute for Medical Knowledge Interchange (IMKI) or the Arden group to create libraries of rules and systems to facilitate their incorporation in various clinical information systems.
The following sections overview several attempts and describe which parts of the vision they either have or are currently focusing on. I also point out the strengths and weaknesses of each approach.
academic research efforts
GLEE is a system for execution of guidelines encoded in the Guideline Interchange Format v3 (GLIF3) (Wang, 2002). GLEE provides an internal event-driven execution model that can be hooked up with the clinical event monitor of a local clinical information system environment. It was developed at Columbia University and is currently only a research prototype. No tools exist for knowledge maintenance other than those proposed by the GLIF3 organization.
GLARE (GuideLine Acquisition, Representation and Execution) is a domain-independent system for the acquisition, representation and execution of clinical guidelines (Terenziani, 2001). GLARE provides expert physicians with an “intelligent” guideline acquisition interface. The interface has different types of checks to provide a consistent guideline: syntactic and semantic tests verify whether or not a guideline is well-formed. GLARE technology has been successfully tested in different clinical domains (bladder cancer, reflux esophagitis, and heart failure), at the Laboratorio di Informatica Clinica, Azienda Ospedaliera S. Giovanni Battista, Torino, Italy. Currently, this application is not available for use by people outside of the research team. See: www. openclinical.org/gmm_glare.html#aimj01
commercial vendors
Therapy Edge (http://www.therapyedge.com/) is a condition-specific, stand-alone, web-based, EMR with extensive clinical decision support for HIV patients. It is a stand-alone tool, that is, it has no integration with the existing CIS; it is only for HIV patients; and has no tools for knowledge based maintenance.
PRODIGY is a guideline-based decision support system in use by a large number of General Practitioners in the United Kingdom (Purves, 1999). The evaluation of PRODIGY 3 is currently underway and Phase 4 is being planned. PRODIGY I and PRODIGY II were implemented as extensions to proprietary UK electronic patient record systems. The PRODIGY system includes a proprietary guideline model, which in PRODIGY II was used to implement guidelines for the management of acute diseases.
PREDICT is a decision support system for delivering evidence-based care directly to clinical practitioners desktops or handhelds. PREDICT integrates with existing clinical information systems using standard internet communication protocols, and allows clinicians to create new disease modules and manage their own guideline content. See: http://www.enigma.co.nz/framed_index.cfm?fuseaction=ourknowledge_products for more info.
eTG complete is an easy-to-use, HTML-based product, available for use on stand alone or networked PC or Mac computers. It covers over 2000 clinical topics and will be updated at regular intervals (three to four times per year). They do not even mention the possibility of integrating their work with any commercially available EMR products. See: http://www.tg.com.au/complete/tgc.htm for an online demonstration. This is just one of many examples of stand-alone, proprietary format, internet-based, guideline presentation systems. In my opinion, none of these are significantly better than what the USA’s National Guideline Clearinghouse (www.guideline.gov) or the New Zealand Guidelines Group (www.nzgg.org.nz) are doing by simply creating a large database of clinical guidelines that are arranged by clinical condition.
AREZZO is a decision support technology (based on PROforma), for building and running clinical applications. AREZZO applications are designed to provide patient-specific advice, guiding the user through data collection, clinical actions and decision making. Applications can be quickly modeled and tested, using the AREZZO Composer, and instantly deployed on the Internet. AREZZO is capable of supporting the development of multi-disciplinary care pathways (customized for local circumstances); its safety-critical and task-based features support the rigor needed for clinical protocols. Currently the system is limited to supporting decisions on pain control in cancer, but this will be expanded in the future to include pain control in much more general terms (for example, arthritis, chronic pain, etc.) See: http://www.infermed.com/ds_arnocs.htm for more information.
summary and conclusions
Library-type applications that provide users with easy access to the latest medical knowledge in the form of journal articles or textbooks will not be going away in the near future. Even with our increasing understanding of how to build intelligent computer systems that can recognize the current patient context and suggest potentially relevant clinical information resources, the users must still have a means of getting directly to the knowledge resources so they can look up additional information. Therefore, work based on the Medline Button, as described by Cimino (1996) must continue. In fact, an effort is now underway within the HL-7 Clinical Decision Support Technical Committee to develop a standard interface that will facilitate further development of these types of applications.
The use and utility of real-time clinical decision support systems (RCDSS) will continue to grow exponentially. As more and more pressure is placed on clinicians to practice evidenced-based medicine and do it as inexpensively as possible, these RCDSSs will proliferate. A recent publication by the Health Information Management Systems Society (HIMSS) entitled, the “Clinical Decision Support Implementers’ Workbook” is now freely available on the internet (Osheroff, 2004). Although little work is currently underway on the development of new hybrid systems, I believe that these types of systems will soon reappear and provide users with significant additional functionality that cannot be achieved using any other means. Finally, as healthcare delivery systems choose and implement commercially available clinical information systems, clinicians and researchers alike could gain valuable knowledge by beginning work on various clinical knowledge management projects with one or more of the vendors or research groups listed above. At the present time, this field has no clear-cut winner. In fact, no system is beyond the advanced research prototype stage. As such, there will be considerable changes in whichever solution is ultimately successful. I believe it is in everyone’s best interest to be a part of the effort in defining and experimenting with these types of systems. Only then will we truly understand what is really useful and how we should proceed.