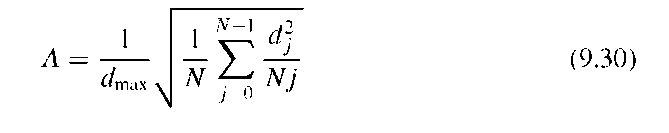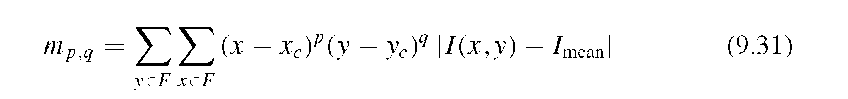The methods described in this topic are widely used in the analysis of biomedical images and computer-aided radiology. Even simple shape metrics that are obtained relatively easily are helpful for diagnosis. Pohlmann et al.29 analyzed x-ray mammograms and used several metrics, including roughness, compactness [Equation (9.4)], circularity [inverse irregularity, Equation (9.6)], the boundary moments [a metric related but not identical to Equation (9.10)], and the fractal dimension of the boundary, to distinguish between benign and malignant lesions. ROC analysis showed the boundary roughness metric to be most successful, followed by a metric of margin fluctuation: The distance between the shape outline and a filtered outline (obtained, for example, by lowpass-filtering the unrolled boundary in Figure 9.10A and by restoring the shape from the smoothed outline function). Boundary moments and fractal dimension showed approximately the same discriminatory power, followed by irregularity and compactness. Pohlmann et al.29 also demonstrated that the spatial resolution of the digitized x-ray images had a strong impact on the ROC curve. A 0.2-mm pixel size lead to poor discrimination, and increasing the resolution to 0.05 mm significantly increased the area under the curve. Further increasing the resolution to 0.012 mm did not significantly increase the area under the curve.
The influence of resolution on the classification of mammographic lesions was also examined by Bruce and Kallergi,7 in addition to the influence of the segmentation method. In this study, both spatial resolution and pixel discretization were changed, and x-rays digitized at 0.22 mm resolution and 8 bits/pixel were compared with x-rays digitized at 0.18 mm resolution and 16 bits/pixel. Segmentation involved adaptive thresholding, Markov random field classification of the pixels, and a binary decision tree. Shape features included compactness, normalized mean deviation from the average radius [Equation (9.7)], and a roughness index. Furthermore, the sequence of rk was decomposed using the wavelet transform and the energy at each decomposition level computed, leading to a 10-element feature vector. Although the wavelet-based energy metric outperformed the other classification metrics, it was noted that the lower-resolution decomposition provided better classification results with the lower-resolution images, an unexpected result attributed to the design of the segmentation process. This study concludes that any classification depends strongly on the type of data set for which it was developed.
A detailed description of the image analysis steps to quantify morphological changes in microscope images of cell lines can be found in a publication by Metzler et al.26 Phase-contrast images were first subjected to a thresholding segmentation, where both the window size and the threshold level were adaptive. The window size was determined by background homogeneity, and the threshold was computed by Otsu’s method within the window. A multiscale morphological opening filter, acting on the binarized image, was responsible for separating the cells. Finally, the compactness, here formulated similar to equation (9.5), characterized the shape. Ethanol and toxic polymers were used to change the shape of fibroblasts, and the compactness metric increased significantly with increased levels of exposure to toxic substances, indicating a more circular shape after exposure.
Often, spatial-domain boundary descriptors and Fourier descriptors are used jointly in the classification process. Shen et al.37 compared the shape’s compactness [Equation (9.4)], statistical moments [Equation (9.13)], boundary moments [Equation (9.11)], and the Fourier-based form factor [Equation (9.25)] in their ability to distinguish between different synthesized shapes and mammographic lesions. Multidimensional feature vectors proved to be the best classifiers, but the authors of this study concluded that inclusion of different x-ray projections and additional parameters, such as size and texture, should be included in the mammographic analysis system. In fact, shape and texture parameters are often combined for classification. Kim et al.21 define a jag counter (jag points are points of high curvature along the boundary) and combine the jag count with compactness and acutance to differentiate benign and malignant masses in breast ultrasound images. Acutance is a measure of the gradient strength normal to the shape boundary. For each boundary pixel j, a gradient strength dj is computed by sampling Nj points normal to the contour in pixel j,
where the f(i) are pixels inside the tumor boundary and the b(i) are pixels outside the boundary. The acutance A is computed by averaging the dj along the boundary:
Therefore, the acutance is a hybrid metric that, unlike all metrics introduced in this topic, includes intensity variations along the boundary. In this study, the jag count showed the highest sensitivity and specificity, followed by acutance and compactness.
A similar approach was pursued by Rangayyan et al.32 Image analysis was performed in x-ray mammograms with the goal of classifying the lesions. The feature vector was composed of the acutance, compactness, the Fourier-domain form factor [Equation (9.25)], and another hybrid metric, a modification of the statistical invariants presented in Equation (9.13). Here, the invariants included intensity deviations from the mean intensity Imean as defined by
In this study, compactness performed best among the single features, and a combination feature vector further improved the classification.
Adams et al.1 found that a combination of a compactness measure and a spatial frequency analysis of the lesion boundary makes it possible to distinguish between fibroadenomas, cysts, and carcinomas, but to allow statistical separation, three MR sequences (T1 contrast, T2 contrast, and a fat-suppressing sequence) were needed. More recently, Nie et al.27 combined shape features (including compactness and radial-length entropy) with volume and gray-level metrics (entropy, homogeneity, and sum average). A neural network was trained to classify the lesion and distinguish between benign and malignant cases.
Fourier descriptors, apart from describing shape features, can be useful for various image processing steps. Wang et al.39 suggested using Fourier coefficients to interpolate the contour of the prostrate gland in magnetic resonance images. Interpolation was performed by zero padding, adding a number of higher-frequency coefficients with complex-zero value. The inverse Fourier transform now has more sample points along the contour. With this technique, contour points from coronal and axial MR scans were joined, and a sufficient number of vertices on the three-dimensional contour was provided for three-dimensional visualization and determination of centroid and size. Lee et al.23 used Fourier coefficients to obtain a similarity measure between outlines of vertebrae in x-ray projection images. In the x-ray image, the vertebral contour was sampled manually, and an iterative curve evolution scheme was used to reduce the number of sample points in their dependence on the local curvature. Based on the remaining sample points, a polygonal interpolation was used to represent the shape. This polygon was then transformed into the Fourier domain, and the root-mean-squared distance between the Fourier coefficients was used to compare contours and provide a similarity metric. Such a similarity metric can also be useful in image retrieval, where similar shapes may occur in different rotational positions or may be scaled. Sanchez-Marln35 used Fourier descriptors to identify cell shapes in segmented edge images of confluent cell layers. The performance of the Fourier descriptors was compared to that of a curvature descriptor. Cell outlines were first filtered to create a smoothed outline and then rotated so that the sequence rk begins with the largest value of rk. In this way, a certain measure of rotation invariance was achieved.
Chain codes have been used frequently to characterize shape features. Shelly et al.36 introduced image processing steps to classify cell nuclei of Pap smear cells as normal or abnormal and to identify extracervical cells. Light microscope images were subjected to edge detection and erosion in preparation for computation of the chain code. The chain code length (in fact, the boundary length) was used to distinguish nuclei from other features in the image. Other features obtained from the chain code were the minimum diameter, the maximum diameter, and the ellipticity. In the same context, that is, images of Pap-stained cervical cells, Bengtsson et al.2 used chain code metrics to detect overlapping cell nuclei. The difference chain code was used primarily to identify pairs of concavities that indicate overlapping convex shapes: in this case, overlapping nuclei. The concavities were then characterized with a number of metrics, such as the spacing along the contour relative to the length of the contour and the minimum distance of the concavities relative to the length of the contour. A decision tree was used to identify the most relevant concavities and therefore to reduce the false positives.
Chain codes were also used for the segmentation of the left ventricular boundaries in cardiac ultrasound images. Zheng et al.44 presented an image processing chain that started with adaptive median filtering for noise reduction and thresholded histogram equalization for contrast enhancement. Edge detection was based on region growing of the low-echogenic center of the ventricle in combination with a gradient threshold to identify edge pixels. The resulting shape was lowpass-filtered, and a polynomial interpolation provided a smoothed contour. Finally, the chain code was obtained and used to determine roundness, area, and wall thickness frame by frame as a function of time.
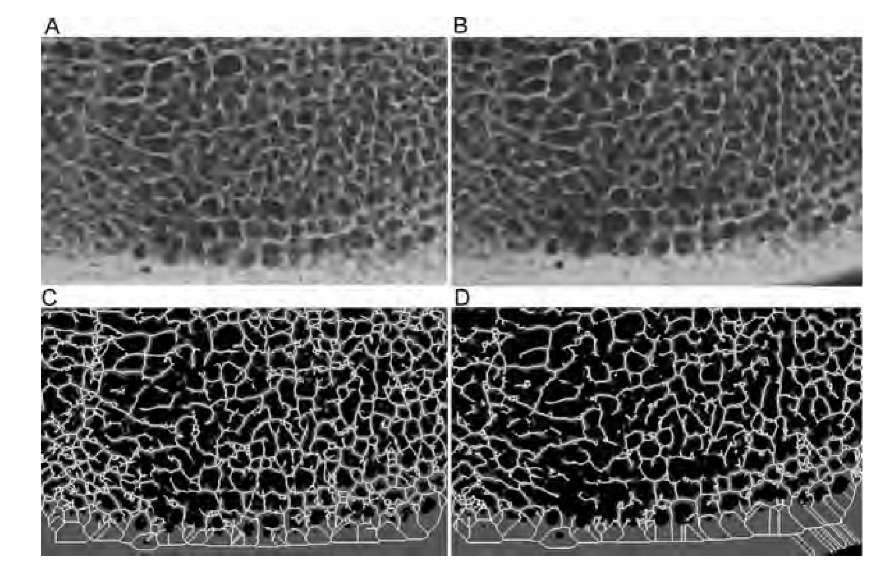
The skeleton of a shape is used when the features in the image are elongated rather than convex and when the features form an interconnected network. A very popular application of connectivity analysis through skeletonization is the analysis of the trabecular network in spongy bone. Spongy bone is a complex three-dimensional network of strutlike structures, the trabeculae. It is hypothesized that bone microarchitecture deteriorates with diseases such as osteoporosis. X-ray projection images, micro-CT images, and high-resolution MR images provide a three-dimensional representation of the trabecular network. An example is given in Figure 9.18, which shows digitized x-ray images of bovine vertebral bone. Figure 9.18A and B show approximately the same region of the same bone, but before the x-ray image in Figure 9.18B was taken, the bone was exposed to nitric acid for 30 min to simulate the microarchitectural deterioration observed in osteoporosis.3 Since x-ray absorption by bone minerals is high, simple thresholding is usually sufficient to separate bone regions from background. Figure 9.18C and D show the thresholded region (gray) and the skeleton (white) of the projected trabecular structure. Visual inspection reveals the loss of structural detail and the loss of connectivity. Skeleton metrics further demonstrate how the connectivity has been reduced by exposure to the acid: There are more isolated elements (33 after acid exposure and 11 before acid exposure), fewer links and holes (2240 links and 725 holes after acid exposure; 2853 links and 958 holes before acid exposure), and fewer vertices (1516 vertices after exposure; 1896 vertices before). Conversely, there is almost no change in scaling-dependent metrics such as the average side length. Figure 9.18 also demonstrates the sensitivity of the skeleton toward noise and minor variations in the shape outline. The lower right corner of Figure 9.18D contains a relatively broad area of compact bone. Skeletonization leads to artifactual diagonal loops. These do not occur in Figure 9.18C because the section of compact bone touches the image border. The area outside the image is considered to be background, and consequently, there exists a straight boundary.
FIGURE 9.18 Microradiographs of a slice of bovine bone (A) and the same bone after 30 minutes of exposure to nitric acid (B). Both radiographs were thresholded at the same level (gray areas in parts C and D), and the skeleton, dilated once for better visualization and pruned to a minimum branch length of 10 pixels, superimposed. Although the changes caused by the acid exposure are not easy to detect in the original radiographs, they are well represented by the connectivity metrics of the skeleton.
In a comparative study15 where the relationship of topological metrics and other shape and texture parameters to bone mineral density, age, and diagnosed vertebral fractures were examined, topological parameters exhibited poor relationship with the clinical condition of the patients. Typical texture metrics, such as the run length and fractal dimension, showed better performance. This study seemingly contradicts the successful measurement of topological parameters in x-ray images of bone biopsies8,17 and high-resolution magnetic resonance images.40 The main difference between the studies is spatial resolution. It appears that a meaningful application of topological analysis to trabecular bone requires a high enough resolution to resolve individual trabeculae, whereas lower-resolution methods such as computed tomography call for different classification approaches.15 Given high enough resolution, the topological parameters allow us to characterize the bone in several respects. Kabel et al.20 used slice-by-slice micrographs of human bone biopsies with a spatial resolution of 25 ^m to reconstruct the three-dimensional network of trabeculae. The Euler number, normalized by the bone volume, was compared with the elastic properties of the bone as determined by finite-element simulations. A weak negative correlation was found between the normalized Euler number (used as a metric of connectivity) and the elastic modulus. These results were corroborated in a study by Yeni et al.,41 who examined micro-CT images of bone biopsies and also found a weak negative correlation between the volume-adjusted Euler number and the elastic modulus. A different notion of the Euler number was introduced by Portero et al.30 and defined as the number of holes minus the number of trabeculae connected around the holes. This modified Euler number was found to correlate well with the elastic modulus of calcaneus specimens determined by actual biomechanical strength tests.31
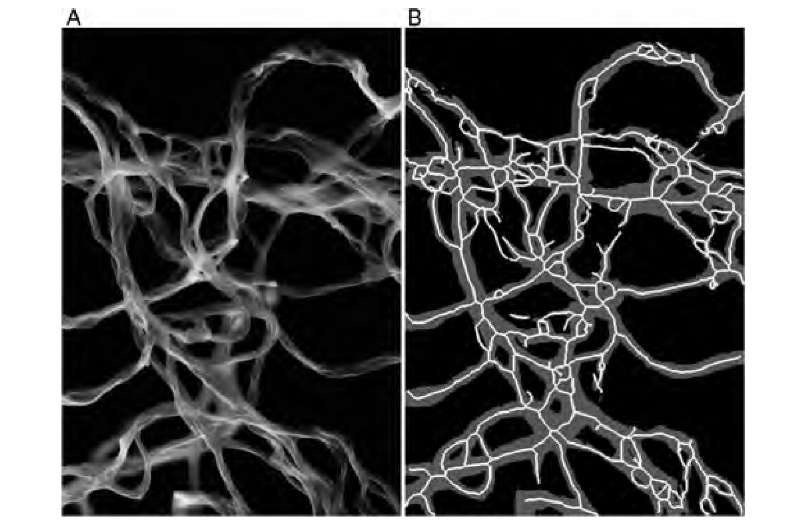
Connectivity was found to be a relevant parameter when a question arose as to whether the presence of gold nanoparticles would influence the cross-linking behavior of collagen fibrils.16 Figure 9.19 shows a representative scanning electron microscopy image of collagen fibrils together with the thresholded and skeletonized binary images. A higher level of cross-linking was related to more curved fibrils, which under visual observation had more self-intersections. Correspondingly, the skeleton showed a higher number of links and a shorter average length of the links. The overall number of holes did not differ between the nanoparticles and the control group, giving rise to a normalized metric of the number of branches relative to the number of holes. In the presence of nanoparticles, this number was found to be 2.8 times larger than in the control group. This ratiometric number was found to be an objective metric of the degree of fibril cross-linking that is independent of the number of fibrils and the microscope magnification and field of view by merit of the normalization.
FIGURE 9.19 Scanning electron microscope image of collagen fibrils (A). After threshold segmentation, the skeleton can be created (B), where the gray region is the thresholded fibril region, and white lines represent the skeleton. Connectivity, represented by the number of edges and the number of nodes, was higher in the presence of gold nanoparticles. The connectivity reflected the tendency of the fibrils to curl and self-intersect when gold nanoparticles were present.