1. Introduction
Genomes range in size from around a million base pairs to many thousands of millions, and yet a typical sequencing reaction yields less than a thousand base pairs of contiguous sequence information. From these tiny fragments of data, the complete genome sequence must be reconstructed as accurately and as completely as possible if genes and other features are to be reliably identified. For the smallest and simplest genomes, the so-called “shotgun” approach is sometimes sufficient: the genome is fragmented and subcloned, and many randomly selected clones are sequenced. As the data accumulates, overlapping stretches of sequence are identified computationally to build contiguous sequences (“contigs”) that grow and merge as the shotgun sequencing project progresses. When enough sequence data has been obtained, the hope is that a single contig can be assembled representing each chromosome.
For more complex genomes, however, shotgun sequencing alone is insufficient. Some parts of the genome may prove refractory to cloning, leaving gaps between a large number of unlinked (and therefore unordered) contigs. Other parts may contain complex repeats – large tracts of sequence that occur at multiple points in the genome – making unambiguous assembly impossible.
Genome maps address this problem by defining the relative positions of selected sequence features over distances of a few kilobases or more, complementing the short-range information provided by the shotgun data. They thus act like the index of a topic, which gives the locations of keywords to help the reader navigate the detailed text. This article will outline the principal methods of genome mapping and will then consider how these approaches are integrated into the sequencing process to produce finished genome sequences.
2. Methods of genome mapping
A number of methods have evolved to produce maps of genomes, but here we will focus only on those methods that give sufficient detail to assist the assembly of sequence data. This largely excludes genetic mapping and fluorescence in situ hybridization, both of which yield coarse maps that are useful mainly for validating the final sequence assembly on the largest scale.
2.1. Physical mapping
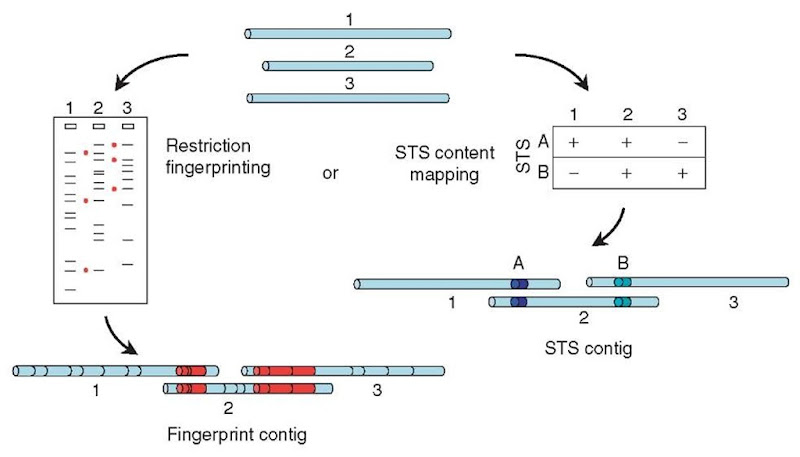
Physical (or clone-based) mapping is conceptually similar to shotgun sequence assembly but takes place on a much larger scale (see Article 13, YAC-STS content mapping, Volume 3 and Article 18, Fingerprint mapping, Volume 3). The aim is to find a series (or “tiling path”) of cloned fragments that represent consecutive overlapping segments of the genome, but the clones that are used are one or two orders of magnitude larger than the small-insert clones used for shotgun sequencing. Typically, the genome is first cloned in BACs (bacterial artificial chromosomes), with insert sizes of 50-200 kb, and each clone in the library is analyzed. This analysis may involve tabulating the sizes of restriction fragments produced when each clone is digested with a chosen restriction enzyme (“fingerprinting”) or testing each of the clones by PCR for the presence of many different specific sequences (known as “sequence tagged sites” or STSs, perhaps chosen from the early data from a shotgun sequencing project). Two clones are then inferred to overlap if they are found to have several restriction-fragment sizes in common or if they both contain the same STS (Figure 1).
Physical mapping suffers from the same limitations as the finer-scale shotgun sequence assembly: regions of the genome that cannot be cloned in BACs leave gaps in the physical map, while very large repetitive tracts (larger than the size of a single BAC clone) can lead to ambiguities in the construction of the tiling path. These problems can be partly overcome in some cases by the use of yeast artificial chromosomes (YACs) as the cloning vector, as these will accept larger inserts and will propagate some sequences that are unstable in bacterial cloning systems. However, YACs are technically more difficult to work with and are also more susceptible to a range of cloning artifacts. In spite of these difficulties, physical mapping is widely used in genome sequencing, often in conjunction with other approaches.
2.2. Optical mapping
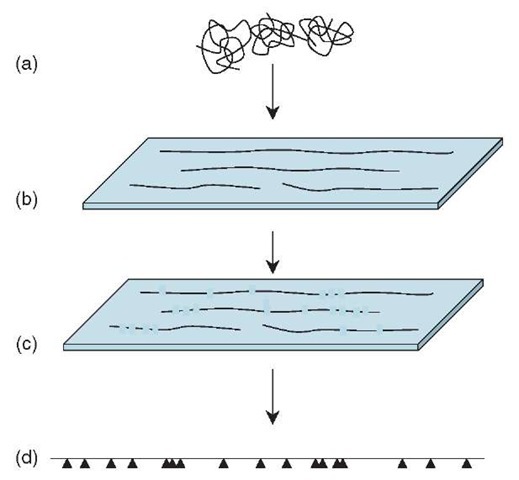
Optical mapping (Samad etal., 1995; Zhou etal., 2003) is an elegant method, though it has so far been applied only to a rather narrow range of genomes. Genomic DNA is spread across a glass slide in such a way that the long molecules are stretched out in one direction and are loosely bound to the surface. The DNA is stained with a fluorescent dye and then treated with a restriction enzyme that cleaves the DNA at each occurrence of the enzyme’s recognition sequence. When examined through a fluorescence microscope, the long DNA molecules appear as dotted lines, broken at each restriction site, and the sizes of the restriction fragments can be measured directly. By compiling many such images, a fairly precise “restriction map” is generated, showing the location of all the restriction sites in the genome. The pattern of restriction sites in the contigs produced by a shotgun sequencing project can then be compared with the genome-wide restriction map to find the precise location of each contig in the genome (Figure 2).
Figure 1 Physical mapping. Large-insert clones (top) can be characterized by digesting them with a restriction enzyme and measuring the sizes of the resulting fragments on a gel (left). Clones that have several fragment sizes in common (indicated by red dots) can be assumed to overlap, with the shared fragments arising from the regions of overlap (bottom left; common restriction fragments shown in red). Alternatively (right), each clone can be tested for the presence of many different STS markers (in this case, two markers A and B). Clones that carry the same STS markers can be inferred to overlap, the shared markers lying in the region of overlap (bottom; markers indicated by colored segments)
The biggest advantage of optical mapping is its independence from cloning and the associated artifacts, though the data it produces (locations of restriction sites) is not as rich as that produced by other methods, complicating its direct use in large genomes.
2.3. Radiation hybrid mapping
Radiation hybrid (RH) mapping is conceptually similar to genetic linkage mapping and gives information on the order and spacing of selected sequences (STSs) in the genome (Cox et al., 1990; see also Article 14, The construction and use of radiation hybrid maps in genomic research, Volume 3). Living cells of the species to be mapped (the “donor”) are irradiated to break their chromosomes at random locations and are then fused with unirradiated cells of another species (the “host”). The result is a population of hybrid cells, each containing the host chromosomes along with a few random fragments of the donor genome. A library of many such cells is then tested, by PCR, to determine which donor-derived STSs are present in each hybrid. If two STSs are close to one another in the donor genome, then they will often remain on the same chromosome fragment after irradiation and hence will often be found together (or “cosegregate”) in the same hybrid cell. Conversely, STSs lying far apart in the donor genome will reside on different fragments after the irradiation and hence will segregate independently amongst the hybrids. By analyzing cosegregation frequencies, therefore, distances between STS markers can be estimated and a map constructed. The basic principle is similar to that used in HAPPY mapping (see below), as illustrated in Figure 3.
Figure 2 Optical mapping. Large fragments of genomic DNA (a) are spread and aligned on a glass surface (b) and stained for visualization. A restriction enzyme cuts the molecules at its recognition sites, and the cuts can be seen using a fluorescence microscope (c). Computerized measurement and analysis of many such images covering overlapping parts of the genome allows the precise pattern (d) of restriction sites (indicated by arrowheads) in the genome to be determined
RH mapping is a powerful tool used to making maps of large genomes since it can be used to map widely spaced markers. However, it suffers from some technical limitations, including the difficulty of making hybrid cells from many donor species, the complicating presence of the host genome in the hybrids, and artifacts caused by biological factors influencing the retention of donor fragments in the hybrid cells.
2.4. HAPPY mapping
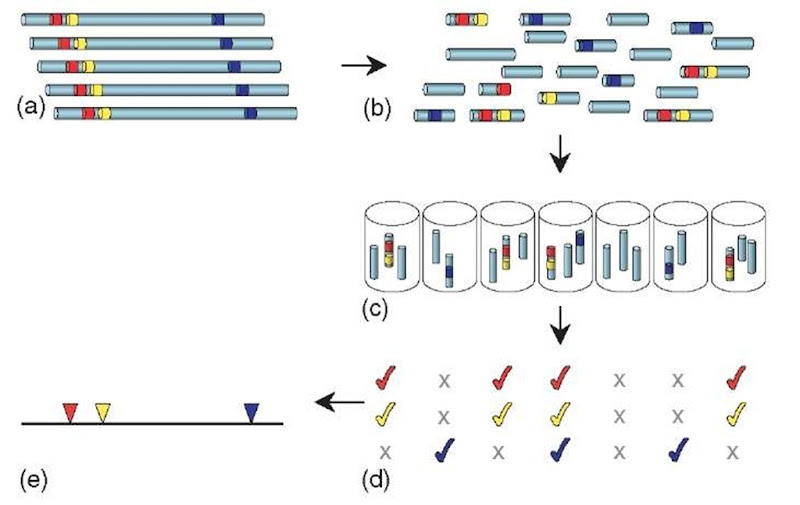
HAPPY mapping (Dear and Cook, 1993; see also Article 22, The Happy mapping approach, Volume 3) is analogous to RH mapping, but is entirely an in vitro process (Figure 3). Again, it begins by randomly breaking genomic DNA of the species to be mapped, either by mechanical means or by radiation. However, instead of segregating the fragments into hybrid cells, they are segregated simply by diluting and dispensing them into a series of samples, each containing only a few random fragments of the genome. Each sample is then screened by PCR to determine the specific sequences (STSs) it contains. Much as in RH mapping, cosegregation frequencies can be used to deduce the order and spacing of the markers.
Figure 3 HAPPY mapping. Genomic DNA (a; colored segments represent STS markers) is broken into random fragments (b), which are greatly diluted and dispensed into a series of samples (c). Each sample is tested by PCR to determine the markers it contains (d). Closely linked markers (red and yellow) will tend to occur together (cosegregate) amongst the samples. By analyzing the cosegregation of many such markers, their order and spacing along the chromosome can be calculated to produce a map (e). The process of radiation hybrid mapping is similar, except that the DNA fragments are propagated in hybrid cells rather than as in vitro samples
The main limitation of HAPPY mapping is the technical challenge of preparing and analyzing minuscule DNA samples, although this is now relatively straightforward. Its advantages arise from its in vitro nature and the consequent control that this gives. The method works equally well on all genomes and does not suffer from biological artifacts induced by specific sequences. Moreover, by choosing how finely to break the DNA at the outset, the level of detail in the maps can be precisely controlled, from relatively coarse long-range maps suited to large genomes (Dear et al., 1998) to detailed maps with accuracies of a few kilobases (Bankier et al., 2003).
3. Mapping as part of a sequencing program
Very few genome sequencing programs use a single methodology in its pure form, but we can outline the elements of two very different, simplified strategies as examples.
In the “top-down” approach, a physical map consisting of overlapping large-insert clones (typically BACs) is first constructed and carefully checked. Some years ago, physical mapping was conducted exclusively using the fingerprinting approach. Nowadays, it is often done by STS content mapping, and, in many cases, a proportion of the STSs would have been previously mapped by RH or other methods to provide an additional layer of positional information and to guard against errors in the physical map. Once the physical map is complete, a set of mapped clones is chosen that covers the genome with the minimum of overlap. Each of the BAC clones in this “minimal tiling path” is then purified, subcloned as small fragments, and sequenced as an individual shotgun project. The genome is therefore sequenced segment by segment in an orderly manner.
The “bottom-up” approach, conversely, starts with extensive whole-genome shotgun sequencing and assembling of the data into sequence contigs. The contigs typically grow and merge as more data are accumulated, until a point of diminishing returns is reached where new sequence data largely duplicates those that are already obtained. At this point, the genome consists of many contigs, separated either by difficult-to-clone regions or by repeats that impede further unambiguous assembly. Mapping strategies are then used to find the arrangement of these contigs in the genome. For example, short sequences chosen from one end of each contig can be HAPPY mapped, showing how the sequence contigs are arranged in the genome. This is often sufficient to resolve repeat-induced gaps; clone-gaps can be closed, for example, by PCR between the sequences now known to flank the gap.
In reality, however, most genome projects use a mixed strategy to bring the sequence to completion. For example, the publicly funded human genome project began as a “top-down” approach, starting with detailed BAC physical maps, reinforced by RH and other marker-based maps. In response to a commercially funded pure shotgun approach, it then adopted a more shotgun-dependent strategy to allow rapid production of an unfinished draft sequence. The public consortium has since largely finished the sequence through a return to more methodical map-led approaches, whereas the pure shotgun strategy of Celera left the genome in over 100 000 unlinked contigs, as expected for a sequence of this size and complexity.
Most large-genome sequencing projects now rely heavily on a combination of shotgun sequencing and physical mapping approaches and also on “map as you go” strategies in which initial sequence data is used to identify those BAC clones that overlap with the existing contigs and those that can be sequenced to extend the contigs in a stepwise manner.
Conversely, the sequencing of the genome of the amoeba Dictyostelium discoideum began as a modified bottom-up approach, with shotgun sequencing not of the whole genome but of individual purified chromosomes. HAPPY mapping was then used to locate each sequence contig precisely in the genome, allowing the gaps in the sequence to be closed methodically using a variety of approaches. This type of approach is widely favored for smaller genomes since the initial shotgun phase yields valuable sequence data, and the subsequent mapping (by whichever means) can be directed toward closing the remaining gaps.