became dominant components of the virus population. Very
was introduced, has never completely recovered and is about
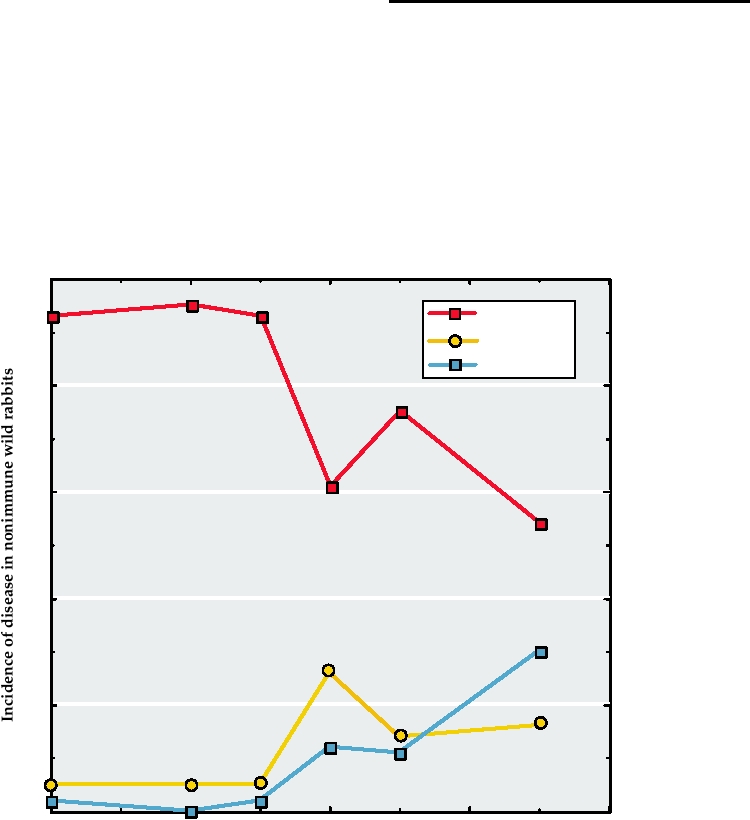
low virulence strains did arise (Fig. 7.9, grade 5) but did
half that today. Efforts are being made to select more viru-
not persist, presumably because they were transmitted less
lent strains of virus that would kill a larger percentage of the
efficiently than were strains of moderate virulence. After a
rabbits. Rabbit calicivirus (Chapter 3) is also being used for
few years, the dominant strains were grades 3 and 4, which
control of rabbits.
kill 5095% of nonimmune, wild-type (i.e., not selected for
The history of myxomatosis in Australia makes clear that
resistance to myxomatosis) European rabbits.
a situation in which a virus rapidly kills the vast majority
At the same time that less virulent virus strains were
of its hosts is inherently unstable. There is selective pres-
being selected, rabbits that were more resistant to myxoma-
sure on the virus to attenuate its virulence and on the host
tosis were also being selected. The enormously high death
to become resistant to the virus. Because both rabbits and
rate caused by viral infection coupled with the short gen-
rabbit myxoma virus multiply rapidly, the accommodation
eration time of rabbits rapidly led to the selection of rabbits
of the virus and host occurred rapidly.
that exhibited increased resistance to the disease caused by
the virus, as illustrated in Fig. 7.10. Perhaps this is the rea-
FAMILY HERPESVIRIDAE
son that strains of low virulence first arose and then faded
from the virus population. Notice that after seven epidem-
ics of myxomatosis, virus strains of grade 3, the dominant
There are more than 100 known herpesviruses which
strain in the virus population, now caused severe disease
are currently classified into three subfamilies called alpha,
in less than 60% of selected rabbits, compared with 95%
beta, and gamma. All but one of the known viruses infect
of unselected (wild type) rabbits. With continuing passage
vertebrates. A partial listing of these viruses is given in Table
of time, the virus and rabbit populations evolved such that
7.7, together with their hosts and the diseases they cause.
rabbit myxoma virus was approximately as serious for the
Most known herpesviruses infect mammals or birds, but
rabbit population as smallpox was for man, that is, about
reptilian, amphibian, and fish herpesviruses also exist. One
40% mortality following viral infection. Nevertheless, the
invertebrate virus, of oysters, has been characterized. The
rabbit population, about 600 million before myxoma virus
viral genome is large, 120230 kb, and the viruses encode
100
% severe
% moderate
% mild
80
60
40
20
0
0
2
4
6
8
Number of epidemics of myxomatosis
FIGURE 7.10 Incidence and severity of disease in nonimmune wild rabbits experimentally inoculated with a strain of
myxomatosis of virulence grade 3 (which induces 7095% mortality in laboratory rabbits) following a given number of
epidemics of myxomatosis. Data from Fenner (1983).
TABLE 7.7 Herpesviridae a
Subfamily/genus/
Virus name
members
abbreviation
Usual host(s)
Transmission
Disease
Alphaherpesvirinae
Simplexvirus
HHV-1 or HSV-1b
Herpes simplex 1
Humans
Infected cells
Cold sores on face and lips
Herpes simplex 2
HHV-2 or HSV-2
Humans
Infected cells
Genital ulcers
Monkey virus Bc
CeHV-1 or HBV
Monkeys
Saliva
Cold-sore like lesions in macaques,
fatal infection in man
Simian agent 8
CeHV-2 or SA8
Vervet monkeys
Saimiriine herpesvirus 1
SaHV-1
Marmosets
Ateline herpesvirus 1
AtHV-1
Spider monkeys
Bovine herpesvirus 2
BoHV-2
Cattle
Several less well known members that infect monkeys and wallabies are not listed here.
Varicellovirus
Varicella-zoster
HHV-3 or VZV
Humans
Aerosols
Chickenpox, shingles
Pseudorabiesd
SuHV-1 o r PRV
Swine
Equid herpesvirus 1,2,4,8,9
EHV-1,-4 etc.
Horses
Aerosols, contact
Respiratory disease, abortigenic
disease
Bovine herpesvirus 1
BoHV-1
Cattle
Felid herpesvirus 1
FeHV-1
Cats
Several less well known members infecting deer, horses, dogs, goats, and cats are not listed separately here.
Mardivirus
Marek's diseasee
GAHV-2,-3
Chickens
Aerosols, contact
T-cell lymphoma
Meleagrid herpesvirus 1
MEHV-1
Turkeys
Iltovirus
ILTVf
GaHV-1 or ILTV
Chickens
Betaherpesvirinae
Cytomegalovirus
Cytomegalovirus
HHV-5 or CMV
Humans
Saliva, urogenital
Disseminated disease in neonates or
excretions
immunocompromised hosts
leading to CNS involvement,
hearing loss, and fatal pneumonitis
Cercopithecine herpesvirus -5, -8
CeHV-5,-8
Primates
Muromegalovirus
Murid herpesvirus-1,-2
MuHV-1,-2
Mice,rats
Saliva
Roseolovirus
Human herpesvirus 6
HHV-6
Humans
Contact, saliva
Exanthum subitum or sixth disease,
may be associated with chronic
fatigue syndrome and/or multiple
sclerosis
Human herpesvirus 7
HHV-7
Humans
Saliva, urogenital
Unknown
excretions
Gammaherpesvirinae
Lymphocryptovirus
Epstein-Barr
HHV4 or EBV
Humans
Saliva, contact
Infectious mononucleosis, Hodgkin's
lymphoma, Burkitt's lymphoma
Numerous other members that infect marmosets, monkeys, orangutans, and apes are not listed separately here.
(Continues)
TABLE 7.7 (Continued )
Subfamily/genus/
Virus name
members
abbreviation
Usual host(s)
Transmission
Disease
Rhadinovirus
Saimiriine herpesvirus 2
SaHV-2
Squirrel monkeys
Human herpesvirus 8
HHV-8
Humans
Contact
Kaposi's Sarcoma
Ateline herpesvirus 2
AtHV-2
Spider monkeys
Numerous other members that infect equids, mice, sheep, and monkeys are not listed separately here.
a
Herpesviruses are generally worldwide in distribution.
b
Although the newer nomenclature lists an adjective describing the species, followed by "herpesvirus" and a number, for example, human herpesvirus 3 or
HHV-3, many authors still use the former names, "varicella-zoster" or VZV, so both forms are shown for the commoner viruses.
c
Monkey virus B is cercopithecine herpesvirus 1; Simian agent 8 is cercopithecine herpesvirus 2.
d
Suid herpesvirus 1.
e
Gallid herpesviruses 2 and 3.
f
ILTV, infectious laryngotracheitis virus or Gallid herpesvirus 1.
many dozens of proteins, which allows them to finely regulate
sion of the viruses into three major lineages classified as distinct
their life cycle. Virions are enveloped, 100300 nm in size, with
subfamilies as well as the division into the nine genera illustrated.
an icosahedral nucleocapsid (Figs. 2.1, 2.5, and 2.20).
All of the viruses in this figure infect mammals or birds. The
mammalian viruses, including the human viruses, are scattered
among all three subfamilies, whereas the known bird viruses
Classification of Herpesviruses
belong to two genera in the subfamily Alphaherpesvirinae.
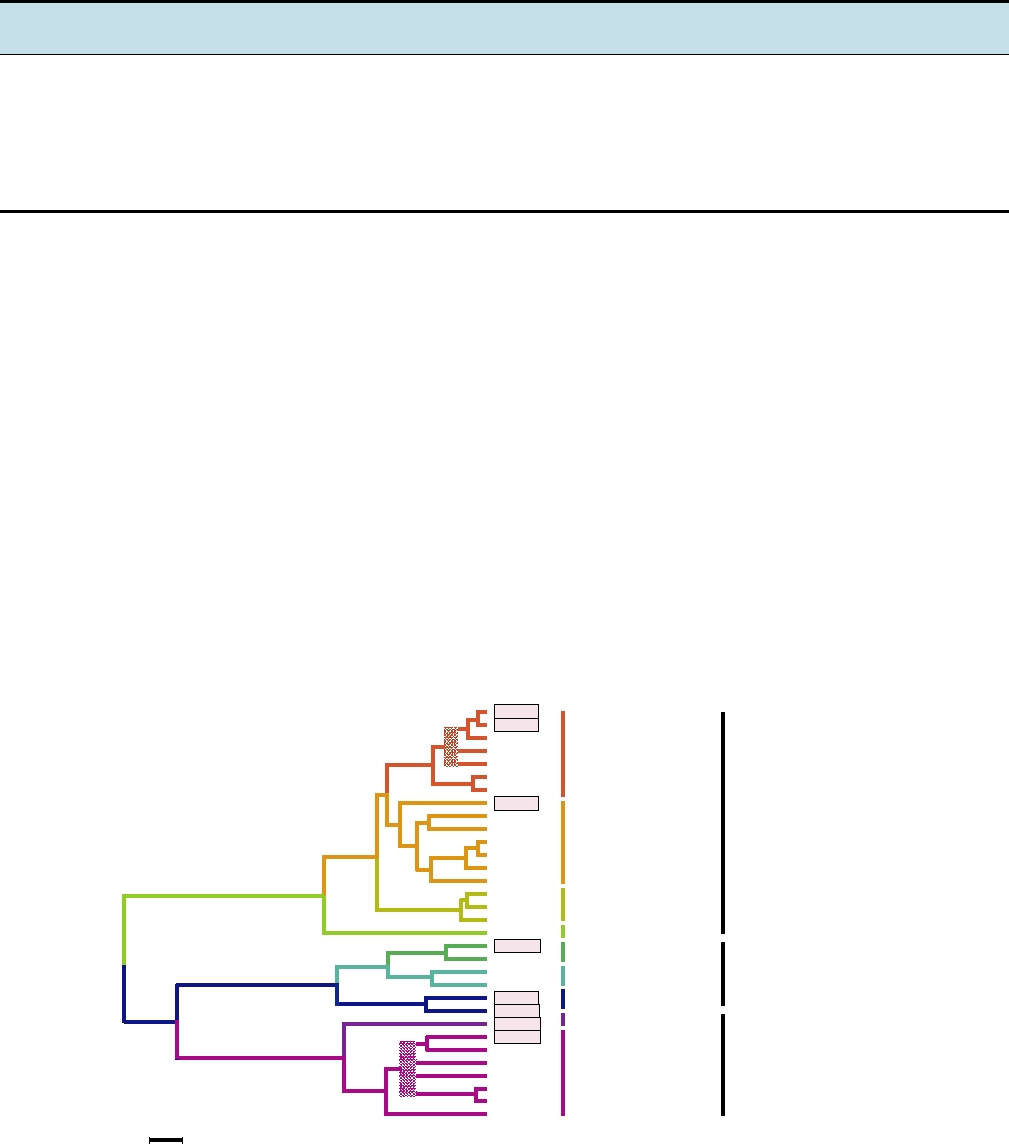
A phylogenetic tree of 32 herpesviruses belonging to nine
Characterized viruses of reptiles also belong to the subfamily
genera is shown in Fig. 7.11. This figure illustrates the divi-
Alphaherpesvirinae. However, viruses of amphibians and
Genera
Subfamilies
HHV-1
HHV-2
CeHV-1
Simplexvirus
MaHV-1
BoHV-2
AtHV-1
SaHV-1
HHV-3
BoHV-1
SuHV-1
Alphaherpesvirinae
Varicellovirus
CaHV-1
PhoHV-1
FeHV-1
EHV-1
GaHV-2
Mardivirus
GaHV-3
MeHV-3
Iltovirus
GaHV-1
Cytomegalovirus
HHV-5
CeHV-8
Muromegalovirus
MuHV-1
Betaherpesvirinae
MuHV-2
Roseolovirus
HHV-6
HHV-7
Lymphocryptovirus
HHV-4
HHV-8
CeHV-17
Gammaherpesvirinae
MuHV-4
Rhadinovirus
BoHV-4
AtHV-4
SaHV-4
EHV-2
0.1 Divergence
FIGURE 7.11
Composite phylogenetic tree for herpes viruses based on amino acid sequence alignments of eight sets
of homologous genes. Most abbreviations are found in Table 7.7. MaHV-1, macropod (wallaby) herpesvirus 1; CaHV-1,
canid (dog) herpesvirus 1; PhoHV-1, phocid (seal) herpesvirus 1. Human herpesviruses are boxed, to emphasize their
distribution throughout the genera. The tree was generated by maximum likelihood; uncertain branches are shown in
heavy patterned lines. Adapted from Fauquet et al. (2005), Figure 5, p. 211.
fish are only distantly related to these three subfamilies and
established in one specific set of cells that are nonpermis-
will probably be classified into a new subfamily in the future.
sive or semipermissive for virus growth and which dif-
Furthermore, the oyster virus is distinct from both the mam-
fer from virus to virus. A different set of cells is lytically
malian viruses and the fish and amphibian viruses and probably
infected so as to produce a progeny virus that is capable of
represents yet another subfamily.
spreading to new hosts. Lytic infection is invariably fatal
Herpesviruses are ancient viruses that have coevolved
for the infected cell. Reactivation of latent virus and lytic
with their hosts. Figure 7.12 illustrates this for several viruses
infection of permissive cells allow the virus to reemerge
in two genera of the Alphaherpesvirinae. Figure 7.12A com-
unchanged and infect new hosts. For some herpesviruses
pares the tree for four simplexviruses with the tree of their
reactivation occurs only sporadically, sometimes only at
host species. The trees are congruent. Similarly, Fig. 7.12B
very long intervals, whereas for other herpesviruses reac-
compares the tree of four varicelloviruses with that of their
tivation occurs more or less continuously and infectious
hosts, and again the trees are congruent. Thus, it is clear that
virus is usually present.
the evolution of these herpesviruses has gone hand-in-hand
The ability of the viruses to latently infect their hosts
with that of their hosts.
for life presents a set of constraints and opportunities for
The tree in Fig. 7.11 also illustrates another interest-
the spread of the virus in nature different from those affect-
ing feature. All of the simplexviruses are primate viruses,
ing most other viruses. The disease caused by the virus
infecting humans and various species of monkeys, except
must be relatively innocuous, or at least not life threaten-
for bovine herpesvirus-2 (BoHV-2). BoHV-2 fits well in
ing, in an immunocompetent native host if lifelong latent
the virus tree, but cattle obviously do not fit in the primate
infection is to be a viable strategy for the virus. However,
tree of Fig. 7.12A. Thus, this bovine virus has not coevolved
the ability of the virus to remain latent and reemerge after
with cattle but appears to have been obtained from a primate.
long intervals means that the virus can persist even if the
An obvious hypothesis is that this represents the spread of
human population is small. Thus, these viruses could have
a virus from humans to their domestic animals, the other
been present in human populations since humans arose,
side of the coin from the transfer of viruses like measles
being passed on from their nonhuman ancestors, as sug-
to humans from domestic animals. Intriguingly, BoHV-2
gested by Fig. 7.12. The interplay between the virus and
causes lesions largely confined to the udders of dairy cat-
the host required to establish lifelong infection in the
tle, which could have come into contact with a human virus
face of a vigorous immune response, described later and
during milking.
in Chapter 10, is further evidence that the herpesviruses
have coevolved with their hosts. Further support for this
idea is the fact that most herpesviruses are worldwide in
Epidemiology of Herpesviruses
distribution. In the case of the human herpesviruses, most
The herpesviruses have a narrow host range and any
are present in all populations of people on earth, including
particular herpesvirus is adapted to use only a single ver-
the most isolated and remote tribes of people that have
tebrate host in nature. All herpesviruses are capable of
been examined.
establishing a latent infection in their natural host whereby
they persist for the life of the animal. Latent infection is
Biology of Herpesviruses
The classification of herpesviruses was originally based
A.
Tree of Some Simplexviruses and Their Hosts
on biological properties, which differ among the three sub-
HHV-1,-2
Human
families (Table 7.8). Although sequence data is now the
preferred means of classification, the subdivision into three
Green Monkey
CeHV-2
subfamilies continues unchanged. Alphaherpesviruses have a
Spider Monkey
AtHV-1
broad host range in the laboratory and will infect a wide vari-
Squirrel Monkey SaHV-1
ety of cultured cells or experimental animals. They spread
rapidly in cultured cells with a short reproductive cycle and
B.
Tree of Some Varicelloviruses and Their Hosts
efficiently destroy the infected cells. In their natural host,
SuHV-1
Pig
latent infections are usually established in sensory neurons
BoHV-1
Cow
and lytic infection often occurs in epidermal cells. The human
EHV
Horse
alphaherpesviruses belong to two genera, Simplexvirus and
Varicellovirus (Table 7.7 and Fig. 7.11). Betaherpesviruses
HHV-3
Human
have a restricted host range and a long infection cycle in
FIGURE 7.12 Evolutionary relationships among the alphaherpesviruses
culture, and infected cells often become enlarged (cyto-
and their hosts. (A) Comparisons of the host and viral trees for several
megaly). In the natural host, the virus is maintained in latent
simplexviruses. (B) Comparison of host and viral trees for several members
form in secretory glands, lymphoreticular cells, kidneys,
of the Varicellovirus genus. Virus abbreviations can be found in Table 7.7.
Adapted from McGeoch et al. (2000).
and other tissues. The human viruses belong to two genera,
TABLE 7.8
Biological Characteristics of the Three Subfamilies of Herpesviruses
Characteristic
Alphaherpesvirinae
Betaherpesvirinae
Gammaherpesvirinae
Host range
Variable, often broad
Restricted
Limited to family of natural host
Reproductive cycle
Short
Long
Relatively long
Infection in cell culture
Spreads rapidly
Progresses slowly
Infects primarily lymphoblastoid cells
Infects many cell types
Cytotoxicity
Much cell destruction
Enlarged cells form
Some lytic infections of epithelial and
fibroblastic cells
Latency
Primarily in sensory ganglia
Maintained in many cells including
Specific for either B or T lymphocytes
secretory glands, lymphoreticular
cells, kidneys, and others
Characteristic genes
Genes in the US sequence and
Genes corresponding to the HHV-5
Genes correspoonding to BNRF-1,
its flanking repeats
US22 family
BTRF-1, and BRLF-1 of HHV-4
Genera
Simplexvirus
Cytomegalovirus
Lymphocryptovirus
Human viruses
HHV-1 (HSV-1)
HHV-5 (CMV)
HHV-4 (EBV)
HHV-2 (HSV-2)
Varicellovirus
Roseolovirus
Rhadinovirus
HHV-3 (VZV)
HHV-6
HHV-8
HHV-7
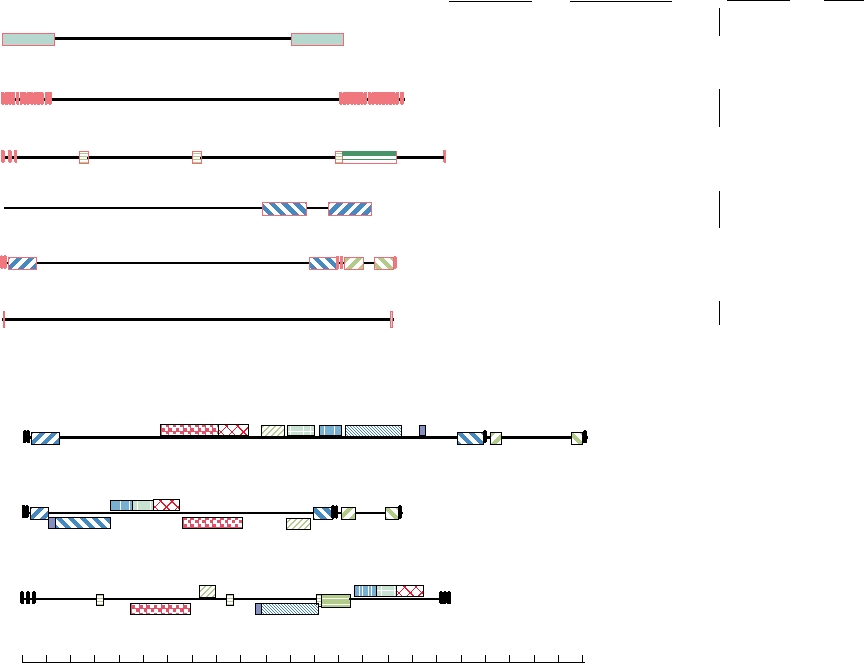
linear sequence of the DNA is fixed (only one isomer exists).
Cytomegalovirus and Roseolovirus. Gammaherpesviruses
The repeated domains in the viruses in Class E of this figure
have the narrowest host range and experimentally infect only
give rise to four isomers; those in Class D give rise to two
members of the family or order to which the natural host
isomers. The genome organizations are important for the
belongs. They replicate in lymphoblastoid cells, and some
expression and replication of the genome, but grouping by
can lytically infect epithelium and fibroblasts. They are spe-
genome organization does not correlate with the taxonomy
cific for B or T cells and infection is frequently latent. There
of herpesviruses based on biological or sequence criteria
are two human gammaherpesviruses, HHV-4 belonging to
(Table 7.7). Thus, the development of the constellation of
genus Lymphocryptovirus and HHV-8 belonging to the genus
repeated sequences may be a more recent occurrence.
Rhadinovirus.
Comparison of the genomes of herpesviruses makes clear
The eight known human herpesviruses can be referred to
that multiple rearrangements have occurred during the evo-
as human herpesviruses (HHV) 1 through 8, but the older
lution of these viruses. This is illustrated in Fig. 7.13B where
names for HHV-1 to 5 are still in common use and are used
the genomes of three human herpesviruses belonging to the
later in this chapter (see Table 7.7). Although they gener-
three different subfamilies are compared. Even though all
ally cause inapparent or innocuous disease, serious illness
share a substantial number of genes, the positions of these
can result, particularly in neonates or in immunocompro-
genes differ in the various genomes.
mised people. Some cancers are also associated with certain
of these herpesviruses. In addition to the human herpesvi-
ruses, at least one herpesvirus of monkeys, Cercopithecine
Structure of the Virion
herpesvirus-1 or B virus, causes a serious, usually fatal ill-
Herpesviruses are enveloped and approximately spheri-
ness of humans that is of concern to animal handlers. These
cal, with a diameter of 100300 nm (Fig. 2.1). They possess
nine viruses and the diseases they cause are described later.
a 100-nm icosahedral nucleocapsid (T=16) that contains at
least six proteins and the viral DNA. The nucleocapsid is
Structure of the Viral Genome
surrounded by or embedded within a structure known as the
tegument (Figs. 2.5 and 2.20). The tegument is composed of
The genomes of herpesviruses are linear dsDNA.
about 20 different virus-encoded proteins and its thickness
Repeated sequence elements are present in most and herpes-
can vary, even within a single virion. Outside the tegument is
virus genomes can grouped into six classes on the basis of the
the envelope containing a dozen or more virus glycoproteins.
location of reiterated domains, as illustrated in Fig. 7.13A.
Many, perhaps all, of the glycoproteins are present in 600 or
In some viruses, including herpes simplex, inverted repeats
more spikes of which several different morphological types
in the DNA lead to inversions of parts of the DNA sequences
can be distinguished. One type of spike is 20 nm long and has
relative to one another during the process of replication.
a globule at its terminus.
These different orientations are called isomers. In others, the
Class
Sequence Arrangement
Virus
Classification
Abbreviation
Common Name
Subfamily
Genus
Ictalurivirus
Channel catfish herpesvirus
IcHV-1
LT
RTR
Unassigned
Unassigned
Green lizard herpesvirus
LaHV-1
A
R
Rhadinovirus
Gamma
Equine cytomegalovirus
EHV-2
Roseolovirus
Beta
HHV6
SaHV-2
Herpesvirus saimiri
B
Kaposi s sarcoma
Rhadinovirus
Gamma
HHV-8
Bovine herpesvirus4
BoHV-4
R1
R4
R3
R2
C
Gamma
HHV-4
Epstein-Barr virus
Lymphocryptovirus
Varicella-zoster
HHV-3
TR
IR
D
Varicellovirus
Alpha
PRV
Pseudorabies
UL
EHV-1,4
Equid herpesviruses
S
anb
b
nc
b
ca
E
HHV-1,-2 Herpes simplex
Simplexvirus
Alpha
Human cytomegalovirus
Cytomegalovirus
Beta
UL
US
HHV-5
LTR
RTR
MCMV-1 Mouse cytomegalovirus
Muromegalovirus
Beta
F
Unassigned
TuHV-1 Tree shrew herpesvirus
Human cytomegalovirus (Betaherpesvirinae, HHV-5)
A
B
CDE
F
G
b
ac
ca
ab
US
UL
Herpes simplex virus (Alphaherpesvirinae, HHV-1)
E DB
G
aa c
ab
ca
US
UL
F
A
U
A
Epstein-Barr virus (Gammaherpesvirinae, HHV-4)
C
EDB
R3
R4
R2R1
G
FC
0
50
100
150
200
kbp
FIGURE 7.13
Upper panel: Grouping of herpesviruses by their genome organization. The narrow lines are the unique
regions of the genomes and the rectangles are repeated domains. These are designated the left and right terminal repeats
(LTR and RTR) in Group A, internal repeats Rl to R4 in Group C, and the internal and terminal repeats (IR and TR) in
Group D. In Group E, both the long and short unique regions are flanked by inverted terminal repeats (shown as ab and
b′a′). In contrast to the LTR and RTR in group A which are almost 10 kbp long, the LTR and RTR in Group F are only
30 bp. Note that this grouping does not correspond exactly with the taxonomy of these viruses as shown in Table 7.7,
which is based on a number of biological characters. Redrawn from Fauquet et al. (2005) p. 195. Lower panel: A more
detailed view of human cytomegalovirus, herpes simplex type 1, and EpsteinBarr virus showing conserved sequence
blocks, which are distinguished by color and pattern. Blocks shown below the midline in HSV and EBV are those in
which the orientation is reversed from that in HHV-5. Redrawn from Gompels et al. (1995). Note that in HHV-5 and
HHV-1 the entire UL region can exist in either of two orientations relative to US.
The nucleocapsid assembles in the nucleus. The mecha-
proteins of enveloped RNA viruses. The tegumented nucleo-
nism by which it is enveloped and released from the cell is
capsid buds into trans-Golgi vesicles, acquiring an envelope,
controversial, and more than one mechanism may be used by
and the completed virion is released from the cell when the
various herpesviruses. One popular model is that the capsid
vesicle fuses with the plasma membrane.
buds through the nuclear membrane into the cytoplasm (Fig.
fection by Herpesviruses
2.25A). The nuclear envelope is a double membrane struc-
ture and the nucleocapsid acquires an envelope upon budding
Many, perhaps all, herpesviruses utilize accessory recep-
through the inner leaflet of the double membrane, then loses it
tors to accelerate virus binding and entry into cells, as
upon fusion with the outer leaflet, resulting in the naked nucle-
described in Chapter 1, and many or all appear to be able
ocapsid being deposited in the cytoplasm. In the cytoplasm the
to utilize more than one high-affinity or entry receptor. In
nucleocapsid acquires the tegument by means of a complex
the case of HSV, glycosaminoglycans serve as accessory
series of proteinprotein interactions. The tegument interacts
receptors and three classes of entry receptors can be used.
with both the nucleocapsid and, during budding, with the viral
These are a member of the tumor necrosis receptor family
glycoproteins, and serves the same function as do the matrix
called herpesvirus entry mediator (HVEM), two cell adhe-
Search WWH :