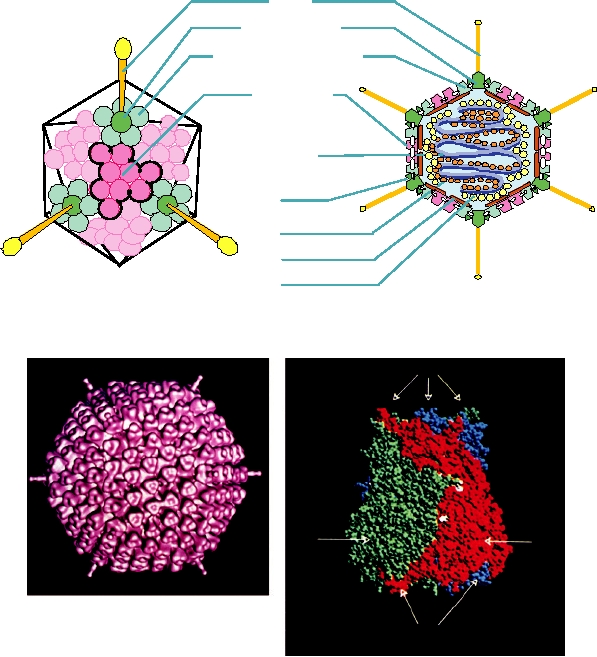
The hexon protein has two eight-strand β sandwiches to
A
give the trimer an approximately sixfold symmetry (and
each hexon protein fills the role of two symmetry units).
There are long loops that intertwine to form a triangular
top. These structures can be fitted uniquely into the enve-
5
lope of density determined by cryoelectron microscopy,
which produces a structure refined to atomic resolution for
A
most of the capsid. The minor proteins can be fitted into
B
3
2
3
this structure.
From the 12 vertices of the icosahedron project long
fibers that are anchored in the surface by a unit called the
penton base. Each fiber terminates in a spherical extension
5
that forms an organ of attachment to a host cell (Fig. 2.12A).
The length of the fiber differs in the different adenoviruses.
B
NONENVELOPED VIRUSES WITH MORE
COMPLICATED STRUCTURAL FEATURES
In addition to the nonenveloped viruses that possess rela-
P
tively straightforward icosahedral symmetry or helical sym-
5
metry, many viruses possess more complicated symmetries
made possible by the utilization of a large number of struc-
tural proteins to form the virion. The tailed bacteriophages
are prominent examples of this (Fig. 2.13). Some of the
3T
2
3
tailed bacteriophages possess a head that is a regular icosa-
S
R Q
hedron (or, in at least one case, an octahedron) connected
to a tail that possesses helical symmetry. Other appendages,
P
such as baseplates, collars, and tail fibers, may be connected
5
to the tail. Other tailed bacteriophages have heads that are
assembled using more complicated patterns. For example,
the T-even bacteriophages have a large head, which can be
thought of as being formed of two hemi-icosahedrons pos-
sessing regular icosahedral symmetry, which are elongated
in the form of a prolate ellipsoid by subunits arranged in a
regular net connecting the two icosahedral ends of the head
FIGURE 2.11 The essential features of the orbivirus native core particle.
of the virus.
The asymmetric unit is indicated by the white lines forming a triangle and
the fivefold, threefold, and twofold axes are marked. (A) The inner capsid
layer of the bluetongue virus (BTV) core is composed of 120 molecules
ENVELOPED VIRUSES
of VP3, arranged in what has been called T=2 symmetry. Note the green
subunit A and the red subunit B which fill the asymmetric unit. (B) The
core surface layer is composed of 780 copies of VP7 arranged as 260
Many animal viruses and some plant viruses are envel-
trimers, with T=13 symmetry. The asymmetric unit contains 13 copies of
oped; that is, they have a lipid-containing envelope surround-
VP7, arranged as five trimers, labeled P, Q, R, S, and T, with each trimer a
ing a nucleocapsid. The lipids are derived from the host cell.
different color. Trimer "T" in blue sits on the icosahedral threefold axis and
Although there is some selectivity and reorganization of lipids
thus contributes only a monomer to the asymmetric unit. From Granoff and
during virus formation, the lipid composition in general mir-
Webster (1999), Color Plate 17.
rors the composition of the cellular membrane from which the
envelope was derived. However, the proteins in the nucleo-
groups of peripentonal hexons are found groups of 9 hexons,
capsid, which may possess either helical or icosahedral sym-
which are sixfold coordinated. Each group of 9 hexons forms
metry, and the proteins in the envelope are encoded in the
the surface of one of the triangular faces. Thus, there are 60
virus. The proteinprotein interactions that are responsible for
peripentonal hexons and 180 hexons in groups of nine.
assembly of the mature enveloped virions differ among the dif-
The structure of the hexon trimer has been solved to
ferent families and the structures of the resulting virions differ.
atomic resolution by X-ray crystallography (Fig. 2.12C).
The virions of alphaviruses, and of flaviviruses, are uniform
A
Fiber
Penton Base
Peripentonal Hexons
Group of 9
Hexons
DNA
IIIa
VII
VI
V
B
C
T
P2
P1
lN
FIGURE 2.12
Structure of adenovirus particles. (A) Schematic drawing of the outer shell of an adenovirus (left), and
a schematic cross section through an adenovirus particle, showing the locations of minor polypeptide components (right).
The virus is composed of 60 peripentonal hexons at the bases of the fibers at the fivefold vertices, and groups of 9 hexons,
one on each triangular face of the icosahedron. (B) Cryoeletron microscopic reconstruction of an adenovirus virion,
viewed down the threefold axis. (C) Space-filling model of the hexon trimer, with each subunit in a different color. The
atomic structure of the hexon has been solved and fitted into the cryoelectron microscopic reconstruction. The locations
of the minor constitutents, indicated schematically in A, were deduced by subtraction. (A) is from Fields et al. (1996)
p. 80, (B) is from Stewart et al. (1991), and (C) is from Athappilly et al. (1994).
structures that possess icosahedral symmetry. Poxviruses,
domain. It binds to the viral RNA and encapsidates it to form
rhabdoviruses, and retroviruses also appear to have a regular
the nucleocapsid. For most RNA viruses, nucleocapsids can
structure, but there is flexibility in the composition of the par-
be recognized as distinct structures within the infected cell
ticle and the mature virions do not possess icosahedral sym-
and can be isolated from virions by treatment with detergents
metry. The herpesvirus nucleocapsid is a regular icosahedral
that dissolve the envelope. The nucleocapsids of alphavi-
structure (Fig. 2.5), but the enveloped herpesvirions are not
ruses, and probably flaviviruses and arteriviruses as well, are
regular. Other enveloped viruses are irregular, often pleio-
regular icosahedral structures, and there are no other proteins
morphic, and are heterogeneous in composition to a greater
within the nucleocapsid other than the nucleocapsid protein.
or lesser extent. The structures of different enveloped viruses
In contrast, the nucleocapsids of all minus-strand viruses are
that illustrate these various points are described next.
helical and contain, in addition to the major nucleocapsid
protein, two or more minor proteins that possess enzymatic
activity. As described, the nucleocapsids of minus-strand
The Nucleocapsid
RNA viruses remain intact within the cell during the entire
The nucleocapsids of enveloped RNA viruses are fairly
infection cycle and serve as machines that make viral RNA.
simple structures that contain only one major structural
The coronaviruses also have helical nucleocapsids, but being
protein, often referred to as the nucleocapsid protein or core
plus-strand RNA viruses they do not need to carry enzymes
protein. This protein is usually quite basic or has a basic
in the virion to initiate infection. The helical nucleocapsids
A. Enterobacteria phage T2
Head
DNA
152 capsomeres
Collar
Neck
Contractile sheath
Fibers
Core
Spikes
Base
plate
Surface view, tail extended
Cutaway view, tail contracted
100 nm
100 nm
Electron micrograph
B. Enterobacteria phage T7
Head
72 Capsomeres
DNA
T=7
Fibers
Tail
100 nm
Surface view
Cutaway view
100 nm
Electron micrograph
C. Lambda-like Phage
Head
72 Capsomeres
T=7
DNA
Tail
Fibers
Surface view
Cutaway view
Electron micrograph
FIGURE 2.13 Morphology of some bacteriophages (members of the Caudovirales). (A) Enterobacteria phage T2,
in the family Myoviridae. The head is an elongated pentagonal structure. (B) Enterobacteria phage T7, a member of the
Podoviridae. (C) Enterobacteria phage λ, a member of the Siphoviridae. All electron micrographs are stained with uranyl
acetate, and all bars shown are 100 nm. Phage diagrams are adapted from Murphy et al. (1995) pp. 51, 60, 55. Electron
micrographs of T2 and T7 were kindly provided by Dr. H.-W. Ackermann, Laval University, Quebec. The electron
micrograph of Ur-Lambda (note the long kinked tail fibers) was kindly provided by Dr. Roger Hendrix.
of (-) RNA viruses appear disordered within the envelope of
lumen of the ER so that they do not aggregate prior to folding.
all viruses except the rhabdoviruses, in which they are coiled
During folding, the solubility of the proteins is increased by
in a regular fashion (see later).
hiding hydrophobic domains within the interior of the protein
The nucleocapsids of retroviruses also appear to be
and leaving hydrophilic domains at the surface.
fairly simple structures. They are formed from one major
The glycoproteins possess a number of important functions
precursor protein, the Gag polyprotein, that is cleaved dur-
in addition to their structural functions. They carry the attach-
ing maturation into four or five components. The precursor
ment domains by which the virus binds to a susceptible cell.
nucleocapsid is spherically symmetric but lacks icosahedral
This activity is thought to be related to the ability of many
symmetry. The mature nucleocapsid produced by cleavage
viruses, nonenveloped as well as enveloped, to bind to and
of Gag may or may not be spherical symmetric. The nucleo-
agglutinate red blood cells, a process called hemagglutina-
capsid also contains minor proteins, produced by cleavage of
tion. The protein possessing hemagglutinating activity is often
GagProPol, as described in Chapter 1. These minor pro-
called the hemagglutinin or HA. The viral glycoproteins also
teins include the protease, RT, RNase H, and integrase that
possess a fusion activity that promotes the fusion of the mem-
are required to cleave the polyprotein precursors, to make a
brane of the virus with a membrane of the cell. The protein
cDNA copy of the viral RNA, and to integrate this cDNA
possessing this activity is sometimes called the fusion protein,
copy into the host chromosome.
or F. The glycoproteins, being external on the virus, are also
The two families of enveloped DNA viruses that we
primary targets of the humoral immune system, in which cir-
consider here, the poxviruses and the herpesviruses, contain
culating antibodies are directed against viruses; many of these
large genomes and complicated virus structures. The nucleo-
are neutralizing antibodies that inactivate the virus.
capsids of herpesviruses are regular icosahedrons but those
The glycoproteins of some enveloped viruses also contain
of poxviruses are complicated structures containing a core
enzymatic activities. Many orthomyxoviruses and paramyxo-
and associated lateral bodies.
viruses possess a neuraminidase that will remove sialic acid
from glycoproteins. The primary receptor for these viruses is
sialic acid. The neuraminidase may allow the virus to pene-
Envelope Glycoproteins
trate through mucus to reach a susceptible cell. It also removes
The external proteins of enveloped virions are virus-
sialic acid from the viral glycoproteins so that these glyco-
encoded proteins that are anchored in the lipid bilayer of the
proteins or the mature virions do not aggregate, and from the
virus or whose precursors are anchored in the lipid bilayer.
surface of an infected cell, thereby preventing released virions
In the vast majority of cases these proteins are glycoproteins,
from binding to it. The viral protein possessing neuraminidase
although examples are known that do not contain bound car-
activity may be called NA, or in the case of a protein that is
bohydrate. These proteins are translated from viral mRNAs
both a neuraminidase and hemagglutinin, HN.
and transported by the usual cellular processes to reach the
The structure of most enveloped viruses is not as rigor-
membrane at which budding will occur. When budding is at
ously constrained as that of icosahedral virus particles. The
the cell plasma membrane, the glycoproteins are transported
glycoproteins are not required to form an impenetrable shell,
via the Golgi apparatus to the cell surface. Some enveloped
which is instead a function of the lipid bilayer. They appear
viruses mature at intracellular membranes, and in these cases
to tolerate mutations more readily than do proteins that must
the glycoproteins are directed to the appropriate place in the
form a tight icosahedral shell and appear to evolve rapidly in
cell. Both Type I integral membrane proteins, in which the
response to immune pressure. However, the integrity of the
N terminus of the protein is outside the lipid bilayer and the
lipid bilayer is essential for virus infectivity, and enveloped
C terminus is inside the bilayer, and Type II integral mem-
viruses are very sensitive to detergents.
brane proteins, which have the inverse orientation with the C
terminus outside, are known for different viruses. Many viral
Other Structural Proteins in Enveloped
glycoproteins are produced as precursor molecules that are
Viruses
cleaved by cellular proteases during the maturation process.
Following synthesis of viral glycoproteins, during which
In some enveloped viruses, there is a structural protein
they are transported into the lumen of the endoplasmic reticu-
that underlies the lipid envelope but which does not form part
lum (ER) in an unfolded state, they must fold to assume their
of the nucleocapsid. Several families of minus-strand RNA
proper conformation, and assume their proper oxidation state
viruses possess such a protein, called the matrix protein. This
by formation of the correct disulfide bonds. This process often
protein may serve as an adapter between the nucleocapsid and
occurs very quickly, but for some viral glycoproteins it can
the envelope. It may also have regulatory functions in viral
take hours. Folding is often assisted by chaperonins present
RNA replication. The herpesviruses also have proteins under-
in the endoplasmic reticulum. It is believed that at least one
lying the envelope that form a thick layer called the tegument.
function of the carbohydrate chains attached to the protein is
The thickness of the tegument is not uniform within a virion,
to increase the solubility of the unfolded glycoproteins in the
giving rise to some irregularity in its structure. The tegument
proteins perform important functions early after infection of a
microscopy, which has been used to determine the structures
cell by a herpesvirus (see Chapter 7).
of several alphaviruses to 725 Å (Fig. 2.5).
More detailed reconstructions of Sindbis virus and Ross
River virus (RRV) have been derived from a combination
Structure of Alphaviruses
of cryoelectron microscopy of the intact virion and X-ray
The alphaviruses, a genus in the family Togaviridae, are
crystallography of alphavirus structural proteins. A cuta-
exceptional among enveloped RNA viruses in the regular-
way view of RRV at about 25-Å resolution is shown in Fig.
ity of their virions, which are uniform icosahedral particles.
2.14A. The nucleocapsid, shown in red and yellow, has a
Virions of two alphaviruses have been crystallized and the
diameter of 400 Å, and is a regular icosahedron with T=4
crystals are regular enough to diffract to 3040-Å resolu-
symmetry. It is formed from 240 copies of a single species of
tion. Higher resolution has been obtained from cryoelectron
capsid protein of size 30 kDa. Note the fivefold and sixfold
A
C
B
N172
R114
S215
T238
M137
W264
L231
H141
D163
A234
G201
I254
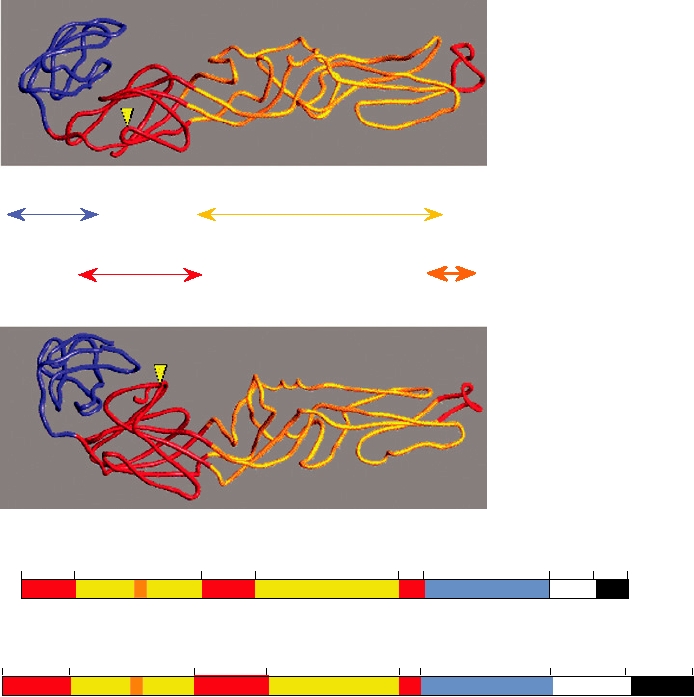
FIGURE 2.14 Structure of Ross River virus reconstructed from cryoelectron microscopy. (A) Cutaway view of the
cryoelectron reconstruction illustrating the multilayered structure of the virion. Envelope glycoproteins are shown in
blue, the lipid bilayer in green, the ordered part of the nucleocapsid in yellow, and the remainder of the nucleocapsid
in orange. (B) Ribbon diagram of the X-ray crystallographic structure of the Sindbis virus capsid protein, with β sheets
represented by large arrows. Only the carboxy-terminal domain, which starts at Arg-114, is ordered in crystals. The
active site residues of the autoprotease, Ser-215, His-141, and Asp-163, are shown in red. The carboxy-terminal Trp-
264, which is the P1 residue of the cleavage site, lies within the active site of the enzyme. The seven residues shown
in yellow-green may interact with the cytoplasmic domain of glycoprotein E2 during budding of the nucleocapsid.
(C) Fit of the Sindbis capsid protein Cα trace (yellow) into the electron density of Ross River virus (blue) determined
by cryoelectron microscopy. (A) and (B) were adapted from Strauss et al. (1995), Figures 4 and 3, respectively, and
(C) was kindly provided by Richard J. Kuhn.
coordinated pedestals in yellow that rise above the red back-
define the structure of the shell of the nucleocapsid to atomic
ground of RNA and unstructured parts of the protein. Each
resolution.
of these pedestals is formed by the ordered domains of one
The envelopes of alphaviruses contain 240 copies of
capsid protein molecule. The lipid bilayer is shown in green
each of two virus-encoded glycoproteins, called E1 and E2.
and is positioned between the capsid and the external shell
E2 is first produced as a precursor called PE2. E1 and PE2
of glycoproteins, shown in blue. The glycoproteins are also
form a heterodimer shortly after synthesis, and both span
icosahedrally arranged with T=4 symmetry. The complete
the lipid bilayer as Type I integral membrane proteins (hav-
structure is therefore quite regular and the virion has been
ing a membrane-spanning anchor at or near the C terminus).
described as composed of two interacting protein shells with
The C-terminal cytoplasmic extension of PE2 interacts in a
a lipid bilayer sandwiched between.
specific fashion with a nucleocapsid protein so that there is
The structure of the ordered part of the capsid protein of
a one-to-one correspondance between a capsid protein and a
Sindbis virus has been solved to atomic resolution by con-
glycoprotein heterodimer. The 240 glycoprotein heterodim-
ventional X-ray crystallography and this structure is shown
ers form a T=4 icosahedral lattice on the surface of the par-
in Fig. 2.14B. The first 113 residues are disordered and
ticle by interacting with one another and with the capsid
the structure is formed by residues 114264. This ordered
proteins. Because of the glycoproteincapsid protein inter-
domain has a structure that is very different from the eight-
actions, the icosahedral lattices of the nucleocapsid and the
fold β sandwich described earlier (compare Fig. 2.14B with
glycoproteins are coordinated.
Figs. 2.3B and 2.4). Instead, its fold resembles that of chy-
At some time during transport of the glycoprotein
motrypsin, and it has an active site that consists of a catalytic
heterodimers to the cell surface, PE2 is cleaved by a cellular
triad whose geometry is identical to that of chymotrypsin.
protease called furin to form E2. E1 and E2 remain associ-
The capsid protein is an active protease that cleaves itself
ated as a heterodimer. If cleavage is prevented, noninfec-
from a polyprotein precursor. After cleavage, the C-terminal
tious particles are produced that contain PE2 and E1.
tryptophan-264 remains in the active site and the enzymatic
In the virion, three glycoprotein heterodimers associate to
activity of the protein is lost.
form a trimeric structure called a spike, easily seen in Figs. 2.5
The interactions between the capsid protein subunits that
and 2.14A. It is not known if the spike assembles during virus
lead to formation of the T=4 icosahedral lattice have been
assembly or if heterodimers trimerize during their transport to
deduced by fitting the electron density of the capsid protein
the cell surface. A reconstruction of a spike of Sindbis virus
at 2.5-Å resolution into the electron density of the nucleo-
at a resolution better than 10 Å is shown in Fig. 2.15A. In this
capsid found by cryoelectron microscopy. Such a recon-
reconstruction, the electron density of E1 has been replaced
struction, based on a cryoEM structure of Sindbis virus at a
by the E1 structure of the related Semliki Forest virus deter-
resolution of better than 10 Å, is shown in Fig. 2.14C. The fit
mined to atomic resolution by X-ray crystallography. The
of the capsid protein is unique and the combined approaches
three copies of E1 project upwards at an angle of about 45°
of X-ray crystallography and cryoelectron microscopy thus
and are shown in three colors because they have slightly dif-
A. Sindbis Virus
B. Dengue Virus
FIGURE 2.15 Comparison of (A) the spike structure of mature Sindbis virus (an alphavirus) with (B) the spike of
immature dengue virus (a flavivirus). The Cα backbones of the three E1 (Sindbis) and E (dengue) glycoprotein ectodomains
are shown in red, green, and blue, as they were fitted into the cryoelectron density envelope. The E1 and E densities have been
zeroed out, leaving the gray envelope that corresponds to E2 for Sindbis and prM for dengue. The density corresponding to
the lipid bilayer is shown in bright green. Adapted from Figure 5 in Y. Zhang et al. (2003) with permission.
ferent environments. The electron density in gray that remains
ers and trimerization of E1 to form homotrimers. The fusion
after subtracting the density due to E1 is thus the electron den-
peptide is exposed and penetrates the target bilayer of the
sity of E2. E2 projects further upward than does E1 and cov-
host endosomal membrane. Fusion follows by methods dis-
ers the apex of E1, which has the fusion peptide. Thus, E2
cussed in Chapter 1.
covers the fusion peptide with a hydrophobic pocket so that it
does not interact with the hydrophilic environment. The apex
Structure of Flaviviruses
of the E2 spike contains the domains that attach to receptors
on a susceptible cell. Both E1 and E2 have C-terminal mem-
Flaviviruses also possess a regular icosahedral structure
brane-spanning anchors that traverse the lipid bilayer shown
(Fig. 2.5) that has been solved by methods similar to those
in green. The C-terminal domain of E1 is not present in the
used to determine the structure of alphaviruses. The struc-
protein whose structure has been determined because hydro-
tures of alphaviruses and flaviviruses are related and have
phobic domains do not easily crystallize. Thus, the electron
descended from a common ancestral structure. Like alphavi-
density shown traversing the lipid bilayer arises from both E1
ruses, flaviviruses produce two structural glycoproteins,
and E2 and shows that the two membrane spanning anchors
called E and prM (for precursor to M). E is homologous to E1
go through as paired α helical structures (Fig. 2.16).
of alphaviruses. Although no sequence identity is detectable,
Upon entry of an alphavirus into a cell, the acidic pH of
the structures of the two proteins are virtually identical and
endosomal vesicles causes disassembly of E2/E1 heterodim-
are formed with a similar fold (Fig. 2.17). prM and E form a
heterodimer and immature particles can be formed if cleavage
of prM is prevented. The glycoprotein heterodimers in these
immature particles trimerize to form spikes whose structure
is very similar to the spikes of alphaviruses (Fig. 2.15B). The
H363
arrangement of the glycoproteins on the immature virus par-
W409
ticle is illustrated in Fig. 2.18A and a cryoEM reconstruction
that illustrates the surface of the immature dengue virus par-
ticle is shown in Fig. 2.18D. The major differences between
I368
the immature flavivirus particle and the alphavirus particle are
that there are 180 copies of the heterodimer in the flavivirus
particle arranged in a T=3 icosahedral structure rather than
F414
240 heterodimers arranged in a T=4 structure in alphaviruses;
that prM is a smaller molecule than PE2 so that in the flavi-
E2
virus spike there is but a thin trace of density that projects
E1
downward, parallel to E, from the cap that shields the fusion
peptide (Fig. 2.15B) rather than a substantial trace of density
M379
in the alphavirus spike (Fig. 2.15A); and that the C-terminal
regions of prM and E that enter the membrane do so inde-
F428
pendently and do not emerge from the internal side of the
membrane (illustrated in Fig. 2.19B), unlike the alphavirus
membrane spanning regions (Fig. 2.16). There is no evidence
that the membrane glycoproteins interact with the nucleocap-
sid in flaviviruses, and the flavivirus nucleocapsid, assuming
it is a regular icosahedral structure, is not coordinated with the
C390
R439
icosahedral structure formed by the spikes, which is again dif-
ferent from alphaviruses where the C-terminal domain of PE2
interacts with the nucleocapsid.
Following cleavage of prM by furin to form M, there is a dra-
matic rearrangement of the flavivirus glycoproteins such that the
T398
final virion structure is very different from the alphavirus structure.
The heterodimers dissociate and E-E homodimers are formed that
collapse over the lipid bilayer (Fig. 2.19). The E-E homodimers
occur in two totally different environments, either perpendicular
FIGURE 2.16 The E1 and E2 transmembrane helices of Sindbis (an
to the twofold axis where they interact with homodimers in side-
alphavirus) determined from a 9Å resolution cryoelectron microscopy
to-side interactions, forming a herringbone pattern, or parallel to
reconstruction. Shown are E1 residues from 409 to 439 and E2 residues
363 to 398 fitted into the transmembrane density. This is Figure 6 from
a twofold axis where they interact at fivefold and threefold axes
Mukhopadhyay et al. (2006), reprinted with permission.
(Fig. 2.18B). A comparison of the immature flavivirus particle,
Semliki Forest E1
C
N
C-Terminal Domain
Dimerization Domain
Central Domain
Fusion Peptide
TBE E Protein
C
N
Semliki Forest E1 Protein
1
39
131
166
289 300
390
413 436
fp
st
tm
TBE E Protein
1
48
138
191
286 301
395
449
496
fp
st
tm
FIGURE 2.17 Comparison of the folds of the Semliki Forest (alphavirus) E1 protein and the tick-borne encephalitis
(flavivirus) E protein. At the top are shown the X-ray crystal structures of the two proteins. At the bottom is a schematic
of the linear amino acid sequences, color coded to indicate which amino acids contribute to each of the domains. Adapted
from Figure 2 in Lescar et al. (2001) with permission.
the mature flavivirus particle, and the alphavirus particle is
Structure of Other Enveloped Viruses with
shown in Fig. 2.18. The mature flavivirus is smooth, with no
Round Nucleocapsids
surface projections, and is 50 nm in diameter (Fig. 2.18E).
The arteriviruses possess icosahedral nucleocapsids, but
The immature particle is ragged in appearance and is 60 nm
the mature virion does not appear to be regular in structure.
in diameter (Fig. 2.18D). The alphavirus virion shows con-
Detailed structures of these particles are not available.
spicuous spikes and is 70 nm in diameter (Fig. 2.18F). The
The herpesviruses are large DNA viruses that have a T=16
arrangement of E or E1 in the three particles is also shown
icosahedral nucleocapsid (Fig. 2.5). A schematic diagram of
(Figs. 2.18A, B, C), illustrating the differences in their
an intact herpesvirion is shown in Fig. 2.20A. Underneath
arrangement.
the envelope is a protein layer called the tegument. The
Entry of flaviviruses follows pathways similar to those
tegument does not have a uniform thickness, and thus the
used by alphaviruses. The acidic pH of the endosome causes
virion is not uniform. Two electron micrographs of herpes-
the E-E homodimers to reorganize to form E homotrim-
virions are shown in Figs. 2.20B and 2.20C that illustrate the
ers. These trimers must reorient so that the exposed fusion
irregularity of the particle and the differing thickness of the
peptide is projected upwards where it penetrates the host
tegument in different particles.
membrane.
100 nm
A
B
C
D
E
F
FIGURE 2.18 Fitting the X-ray crystal structures of flavivirus E protein and alphavirus E1 protein into the respective
cryoelectron density envelopes of the virions. (A) Dengue E protein fitted into the cryoelectron density of immature prM-
containing particle; (B) dengue E protein fitted into the envelope of the mature virion; (C) the fit of Sindbis alphavirus
E1 into the Sindbis virus envelope; (D) cryoelectron reconstruction of the immature dengue prM-containing particle
at 16-Å resolution; (E) cryoelectron reconstruction of the mature dengue virion at 12-Å resolution; (F) cryoelectron
reconstruction of Ross River alphavirus at 25-Å resolution. Panels A and C were provided by Richard J. Kuhn; panel B
is adapted from Figure 3c in Kuhn et al. (2002) with permission; panel D is adapted from Figure 3b in Y. Zhang et al.
(2003) with permission; panel E is reprinted from Figure 1a in W. Zhang et al. (2003) with permission; panel F is adapted
from Figure 4 in Strauss et al. (1995) with permission.
The retroviruses have a nucleocapsid that forms initially
In gammaretroviruses, the capsid forms during budding,
using spherical symmetry principles. Cleavage of Gag during
and the nucleocapsid is round and centrally located in the
virus maturation results in a nucleocapsid that is not icosa-
mature virion. This process is illustrated in Fig. 2.21C for
hedral and that is often eccentrically located in the virion.
murine leukemia virus. The top micrograph shows a bud-
Fig. 2.21A presents a schematic of a retrovirus particle that
ding particle with a partially assembled capsid. The bottom
illustrates the current model for the location of the various
micrograph shows a mature virion.
proteins after cleavage of Gag and GagPol. Figs. 2.21B, C,
In the lentiviruses, of which HIV is a member, the cap-
and D show electron micrographs of budding virus particles
sid also forms as a distinct structure only during budding.
and of mature extracellular virions for three genera of retrovi-
In the top panel of Fig. 2.21D is shown a budding particle
ruses. Betaretrovirus particles usually mature by the formation
of bovine immunodeficiency virus. After cleavage of Gag
of a nucleocapsid within the cytoplasm that then buds through
to form the mature virion, the capsid usually appears cone
the plasma membrane. This process is shown in Fig. 2.21B for
shaped or bar shaped (bottom panel of Fig. 2.21D).
mouse mammary tumor virus. In the top micrograph in Fig.
MasonPfizer monkey virus is a betaretrovirus whose
2.21B, preassembled capsids are seen in the cytoplasm. In the
capsid is cone shaped and centrally located in the mature
middle micrograph, budding of the capsid through the plasma
virion. A single amino acid change in the matrix protein
membrane is illustrated. In the bottom micrograph, a mature
MA determines whether the capsid preassembles and then
virion with an eccentrically located capsid is shown.
buds, or whether the capsids assemble during budding.
A
Immature Virion
Mature Virion
prM(18 kDa)
E dimer
E(~50 kDa)
Membrane
M(8 kDa)
Nucleocapsid
RNA:~11 kb
C protein: 13 kDa
(prM/E)3 heterotrimer
E-E homodimer
B
III
I
II
M-H
CS
1
E-H
E-H2
FIGURE 2.19 (A) Schematic representation of maturation of dengue virus. In the immature particle, as shown in panels
(A) and (D) of Fig. 2.18, three heterodimers of E and prM come together to form a heterotrimer. Upon cleavage of prM to
M and pr, E collapses onto the surface of the mature particle (panels B and E in Fig. 2.18) as homodimers. (B) Diagram
of the dengue virus E protein in the mature particle as derived from cryoEM. This shows the three domains colored in the
same way as in Figs. 2.17 and 2.18 as well as the locations of the transmembrane and intramembrane helices in blue and
orange for E and M, respectively. Reprinted from Figure 4a in W. Zhang et al. (2003) with permission.
100 nm
100 nm
A
B
C
FIGURE 2.20 Two views of herpes simplex virus. (A) Cutaway schematic representation showing the outer envelope
with projecting spikes, the irregular inner margin of the envelope due to the tegument, and the icosahedral core containing
162 capsomeres in a T=16 arrangement. One of the triangular faces of the icosahedron is outlined. Adapted from Murphy
et al. (1995) p. 114. (B) Negatively stained electron micrograph of an intracellular particle of bovine herpesvirus.
(C) Section through a bovine herpesvirion. Images in (B) and (C) were kindly provided by Dr. Peter Wild.
A
B. Betaretrovirus
C. Gammaretrovirus
D. Lentivirus
Genome
gag
env
pol
CAP
An
pro
Pol Proteins
Env Proteins
Gag Proteins
INT
SU
NC
RT
CA
TM
MA
PRO
Lipid Bilayer
RNA
FIGURE 2.21 Structure of retrovirus particles. (A) Schematic cross section through a retrovirus particle. The lipid
bilayer surrounds the particle and has imbedded in it trimeric spikes composed of surface (SU) and transmembrane (TM)
envelope proteins. The internal nonglycosylated proteins are encoded by the gag gene and include NC, the nucleocapsid
protein complexed with the genomic RNA, CA, the major capsid protein, and MA, the matrix protein that lines the inner
surface of the membrane. Other components include RT, the reverse transcriptase, IN, the integrase, and PR, the protease.
Adapted from Coffin et al. (1997), Retroviruses, p. 2. (B) Electron micrographs of mouse mammary tumor virus particles.
Top: intracytoplasmic particles; middle: budding particles; bottom: mature extracytoplasmic particles. (C) Electron
micrographs of murine leukemia virus particles. Top: budding particles; bottom: mature extracytoplasmic particles.
(D) Electron micrographs of bovine immunodeficiency virus. Top: budding particles; bottom: mature extracytoplasmic
particles. Adapted from Coffin et al. (1997), Retroviruses, p. 30.
Thus, the point at which capsids assemble does not reflect
between the nucleocapsid and the glycoproteins. The lack of
a fundamental difference in retroviruses. Preassembly of
such interactions permits these viruses to form pseudotypes,
capsids or assembly during budding appears to depend on
in which glycoproteins from other viruses substitute for
the stability of the capsid in the cell. Stable capsids can pre-
those of the virus in question. Pseudotypes are also formed
assemble. Unstable capsids require interactions with other
by retroviruses.
viral components to form as a recognizable structure.
The structures of paramyxoviruses and orthomyxoviruses
are illustrated schematically in Fig. 2.22. The helical nucleo-
capsids contain a major nucleocapsid protein called N or NP,
Enveloped Viruses with Helical
and the minor proteins P (NS1) and L (PB1, PB2, PA), as shown.
Nucleocapsids
There is a matrix protein M (M1) lining the inside of the lipid
The coronaviruses and the minus-strand RNA viruses
bilayer and also two glycoproteins anchored in the bilayer that
have nucleocapsids with helical symmetry. The structures
form external spikes. The two glycoproteins, called F and HN
of the mature virions are irregular, with the exception of
in paramyxoviruses and HA and NA in orthomyxoviruses,
the rhabdoviruses, and the glycoprotein composition is not
do not form heterodimers but rather form homooligomers so
invariant. Because of the lack of regularity in these viruses,
that there are two different kinds of spikes on the surface of
as well as the lack of symmetry, detailed structural studies of
the virions. HA in the orthomyxoviruses forms homotrim-
virions have not been possible. The lack of regularity arises
ers whereas NA forms homotetramers, and the two types of
in part because in these viruses there is no direct interaction
spikes can be distinguished in the electron microscope if the
A
Paramyxovirus genome
Names of measles virus
N
P
M
F
L
HN
gene products
Symbols
NP
NS1
M1
HA
NA
PB1, PB2, PA
Comparable influenza
virus gene products
C
B
A paramyxovirus
particle
D
Orthomyxovirus
particles
Lipid Bilayer
RNA
FIGURE 2.22 Morphology of orthomyxoviruses and paramyxoviruses. (A) Schematic of the genome organization
of a paramyxovirus, Sendai virus. The names of the gene products and symbols to be used in the diagram are indicated.
Also shown are the comparable gene products of influenza virus, an orthomyxovirus. (B) Schematic cutaway view of
an orthomyxovirus or paramyxovirus particle. The nucleocapsid consists of a helical structure made up of the RNA
complexed with many copies of the nucleocapsid protein. This internal structure also contains a few molecules of the
RNA polymerase L (or PB1, PB2, PA in influenza virus), and P (or NS1). The nucleocapsid is enveloped in a lipid bilayer
derived from the host cell in which are embedded two different glycoproteins, F and HN (or HA and NA in influenza
virus) and which is lined on the inner surface with the matrix protein M. (C) Electron micrograph of a thin section of
a measles particle. This photo was taken by Cynthia S. Goldsmith and obtained from the Public Health Image Library
(PHIL). (D) Electron micrograph of a negatively stained influenza virus particles. This photo was taken by Fred Murphy
and obtained from PHIL.
resolution is high enough. As occurs in many enveloped RNA
The structures of rhabdoviruses and filoviruses are
viruses, F and HA are produced as precursors that are cleaved
illustrated in Fig. 2.23. The rhabdoviruses assemble into
by furin during transport of the proteins. Cleavage is required
bullet-shaped or bacilliform particles in which the helical
to activate the fusion peptide of the virus, which is found at the
nucleocapsid is wound in a regular elongated spiral con-
N terminus of the C-terminal product (see Fig. 1.6). Electron
formation (Figs. 2.23B and C). The virus encodes only five
micrographs of virions are shown in Figs. 2.22C and D. The
proteins (Fig. 2.23A), all of which occur in the virion (Fig.
particles in the preparations shown are round and reasonably
2.23B). The nucleocapsid contains the major nucleocapsid
uniform, but in other preparations the virions are pleomorphic
protein N and the two minor proteins L and NS. The matrix
baglike structures that are not uniform in appearance. In fact,
protein M lines the inner surface of the envelope, and G is an
clinical specimens of some orthomyxoviruses and paramyxo-
external glycoprotein that is anchored in the lipid bilayer of
viruses are often filamentous rather than round, illustrating the
the envelope. Budding is from the plasma membrane (Fig.
flexible nature of the structure of the virion. The micrograph
2.23D).
of the paramyxovirus measles virus shown in Fig. 2.22C is a
The filoviruses are so named because the virion is fila-
thin section and illustrates the lack of higher order structure
mentous. A schematic diagram of a filovirus is shown in Fig.
in the internal helical nucleocapsid. The micrograph of the
2.23E, and electron micrographs of two filoviruses, Marburg
orthomyxovirus influenza A virus shown in Fig. 2.22D is of
virus and Ebola virus, are shown in Figs. 2.23F and G. Notice
a negative-stained preparation and illustrates the spikes that
that in the electron microscope, filovirus virions often take
decorate the virus particle.
the shape of a shephard's crook or the number 6.
A.
Genome organization
N
M
G
L
NS
Viral proteins
Lipid Bilayer
RNA
Other viral components
B.
C.
D.
E.
F.
G.
500 nm
500 nm
FIGURE 2.23 Morphology of the Rhabdoviridae and Filoviridae. (A) Genome organization of vesicular stomatitis
virus (VSV), a Vesiculovirus, with the symbols for the various viral components shown below. (B) Cutaway diagram of
a VSV particle. (C) A negatively stained electron micrograph of VSV virions. (D) Surface replica of a chicken embryo
fibroblast infected with VSV. Note that the magnification is approximately 1/10 of that shown in (C). (E) Diagram
of a filovirus, using the same color code for the components. (F) Negatively stained preparation of Marburg virus.
(G) Filamentous forms of Ebola (Reston) virus. (B) is from Simpson and Hauser (1966); (F) and (G) are from Murphy
et al. (1995) p. 289; and (D) is from Birdwell and Strauss (1975).
Search WWH :