Mutations observed
E-gene mutations
In case of DEN-2 virus, maximum numbers of viral isolates have been analyzed in studies from Thailand. The E-NS1 region of 77 different variants of DEN-2 studied using Maximum Parsimony analysis of 240 nucleotide sequence, showed 11 of 240 nucleotide variation; 4.6% divergence but did not reveal significant segregation of virus according to geographic location (Rico-Hesse R. et al. 1998). Similarly, Phylogenetic analysis of 120 E gene of DEN-2 by another group from Thailand has confirmed existence of six genotypes of this virus; however evolutionary relationships among the genotypes is difficult to determine (Zhang, C. et al 2006). In terms of dengue pathogenesis these studies failed to show segregation of DF versus DHF-associated viruses on the evolutionary tree. There are no clear-cut evolutionary divergence or branching of DF versus DHF isolates, suggesting that nucleotides from this region of the genome encode amino acids that are apparently not under immune selection (Rico-Hesse R. et al. 1998 and Zhang, C. et al 2006, ). DEN-3 has replaced DEN-2 as most frequently isolated virus in Thailand since late 1980′s (Wittke,V.et al.,2002). The evolutionary history of Thai DEN-3 viruses, has been studied by comparative analysis of the nucleotide sequence of E protein genes of currently prevailing isolates with those from all previously published E gene sequences of DEN-3 virus available in Gen Bank (Wittke,V et al. 200218), this study has shown E-gene of DEN-3 to be relatively conserved at amino acid level, however, four amino acid changes have been identified within genotype II of Thai strains. The amino acid changes observed at positions (E172 I-V) and (E479 A-V) are the only difference found between pre and post-1992 viruses. Similarly, there is little evidence to support in-situ evolution among the virus samples that were studied over prolong period ranging from days to months in a selected community in Thailand (Jar man, R.G.et al.,2008) very few mutational changes were noted, and association of these mutations with disease severity could not be delineated either. Analysis by E-sequence of eight DEN-3 strains from Bangladesh (2002 out-break strains) were found to be very closely related to Thai isolates that caused out-break in 1998 in Thailand. The multiple alignment of amino acid (aa) sequence revealed that Bangladeshi isolates and Thai isolates shared common aa changes at position E127 (I-V), suggesting that 2002 outbreak in Bangladesh was due to introduction of Thai isolates (Islam, M.A.et al.,2006), however the statistical association of aa changes with disease severity could not be delineated. In case of Sri Lankan DEN-3, type III is the most frequent strain with two distinct clades IIIA and IIIB linked to mild and severe disease epidemics on the island respectively (Kanakaratne, N. et al.,2009). Phylogenetic studies of E-NS1 junction of DEN-2 isolates from Sri Lanka has categorized the isolates into 4 genotypes designated as Malaysian/Indian subcontinent, Southeast Asian, American, and West African (Sylvatic) and Sri Lankan isolates are closely related to Indian / Malaysian genotype.
C-prM mutations
The phylogenetic analysis of 433 base pair region (nucleotides 180 — 612) of the DEN-3 CprM gene junction showed that sequences of Delhi isolates (2006 outbreak) were closely related to sequences from Guatemala (1998) and presented a nucleotide identity of 95.9 — 98.2% (mean 97.05%). On comparison of Delhi 2006 sequences with other Indian sequences from years 2003, 2004, and 2005, mean sequence divergence of 2.85%, 2.15%, and 1.6%, respectively, were observed (Kukreti, H. et al.,2008. Common amino acid mutations observed in 2006 DENV-3 sequences are given in table 3. Similar study performed on DEN-3 isolates of 2003-04 outbreaks in New Delhi, found them to be closely related and belonged to subtype III from Sri Lanka (Dash, P.K.et al.,2006). Moreover, Phylogenetic analysis of C/PrM/M region of DEN-3 isolates from Pakistan (2004-05 outbreak isolates) also showed sequence homology with 2003-04 New Delhi outbreak strains suggesting that circulation of common isolates of DEN-3 subtype III in the region . There was no clear statistical association of disease severity with specific serotype, as viruses isolated from DHF patients fell at different locations on the phylogenetic tree (Kukreti, H. et al.,2008;Dash, P.K.et al.,2006).
|
DENV-Protein |
Geographical Origin |
year |
aa change (positions) |
Relation to Disease severity |
Ref |
|
Envelope (E) Den 3 |
Thailand |
2002 |
E124 (P-S) E132 (H-T) E172 (I-V) E479 (V-A) |
Could not be ascertained |
(Rico-Hesse R et al) |
|
Envelope (E) Den 3 |
Thailand |
2008 |
Phylogenetic analysis= multiple genetic variants (mutational positions not mentioned) |
Could not be ascertained |
(Jarman RG) |
|
DEN 3 E region |
Taiwan |
2008 |
E301 (L to T) |
(King, C.C. et al.,2008) |
|
|
Envelope (E) Den 3 |
Bangladesh |
2005/6 |
E81 (I-T) E140 (I-T) E127 (l-V) |
Distinct clade causing epidemic outbreak |
(Islam, M.A.et al.,2006) |
|
Den 3 C-preM/E |
Srilanka |
2003-06 |
Phylogenetic analysis= multiple genetic variants (mutational positions not mentioned) |
2 distinct clades linked to mild (IIIA) and severe (IIIB) disease epidemics |
(Kanaka-ratne et al) |
|
CprM Den 3 |
India |
2005/6 |
CprM88 (I-V) CprM 121(A-A) CprM127 (i-P) CprM122 (G-G) CprM55 (A-L) CprM 128(V-G) |
No association of any particular variant with serious dengue disease |
(Kukreti H) |
|
C-prM |
India |
2006 |
C-prM108(M-I) C-prM112(T-A ) |
may be attributed to increased incidence of DHF & DSS in India |
(Dash PK) |
|
CprM Den 3 |
Pakistan |
20052006 |
C-prM |
Similar to New Delhi strain 2004 |
Jamil B, et al |
|
DEN 3 prM region |
Taiwan |
2008 |
CprM 55 (L-H) PrM 57 (T-A) |
No association with disease severity could be determined |
(King CC) |
Table 2. Genetic Characteristics and relation to disease severity in patients with DEN-3 infections reports from South East Asian Region
Using maximum likelihood and Bayesian approaches, phylogenetic analysis of Taiwan’s indigenous DENV-3 isolated from 1994 and 1998 dengue/DHF epidemics were found to be of three different genotypes -I, II and III each associated with DEN-3 circulating in Indonesia, Thailand and Sri Lanka, respectively(King, C.C. et al.,2008). The authors of this study analyzed complete nucleotide sequence of DEN-3 for its mutation and its relation with regional evolution. The highest level of nucleotide sequence diversity, and the positive selection site was detected at position 178 of the NS1 gene. Although the authors have identified the NS 1 gene as the positive selection site and the envelope protein site for purifying selection pressure, however direct association of these changes with disease severity was not determined. Study from Bangkok Thailand performed sequence analysis on E/NS-1 region of Thai isolates to determine if viral strains from less severe DENV infections had distinct evolutionary nucleotide pattern then those with more severe form (Rico-Hesse R. et al.,1998). This study found that two distinct genotypes were identifiable from both DF and DHF cases, suggesting its evolution from common progenitor that perhaps shares the potential to cause severe disease.
|
DENV-Protein |
Geographical Origin |
year |
aa change (positions) |
Relation to Disease severity |
Ref |
|
E/NS1 (77 DEN-2 virus strains studied) |
Thailand |
1998 from 1980 |
11 nucleotides (4.6% divergence) between Strain PU0-218-280. 22 nt or 9.2% divergence PUO-218-D80141 |
No specific association with disease severity |
(Klungthong C) |
|
E/NS1 junction Den 2 |
Srilanka |
2003-06 |
239-nt (from positions 2311-2550) |
Could not be ascertained |
(Kanaka-ratne et al) |
|
3′ and 5′ UTR Den 2 |
Thailand |
1996-97 |
5′ NCR homologus 3′ UTR trinucleotide change 297± 299 (two transversions and one transition) |
Trinucleotide change may alter the functional characteristic of Secondary structure |
(Mangada MNM) |
|
Den 2 E /C/NS2A |
Thailand |
2006 |
approx 10-3 substitutions |
no apparent association |
(Zhang C) |
|
3′-UTR |
Thailand |
1973 to 2003 |
Variable secondary structures were detected |
No clear association |
(Zhou Y) |
Table 3. Genetic Characteristics and relation to disease severity in patients with DEN-3 infections reports from South East Asian Region
Untranslated Region (UTR) mutations
The 3′ UTR region is thought to play a pivotal role in the DENV biology; it contains several conserved regions as well as 3′ long Stable Hair Pin structure which is conserved among all the members of the family Flaviviridae. It has been proposed that this structure interacts with viral and host nucleic acid and protein factors to form a complex to regulate transcription and replication (Zhou, Y. et al.,2006. Therefore it appears to play a significant role in the efficiency of RNA- translation, and virus ability to cause infection, hence the role of 3-UTR in determining the severity of dengue disease seems plausible. The literature reviewed under this study did show considerable intra-serotype diversity at 3-UTR region with greatest variability seen in DEN-4 followed by DEN-1.
A comparative analysis of 3′ UTR conducted for DENV isolates from Bangkok, Thailand compared Thai sequences with 61 globally sampled isolates of DENV taken from patients with varying disease severity. Although some genetic variations were found both within and among the serotypes notably at 3′ Long Hairpin Stable structure, however these mutations did not show consistent association with the clinical outcome of the DENV infection (Zhou, Y. et al.,2006). Study focusing on terminal 3′ 5′UTR sequences of four DEN-2 from Thailand 1998 outbreak strains, showed complete homology for sequences at 5′ UTR (highly conserved region) when compared with the prototype virus New Guinea C strain
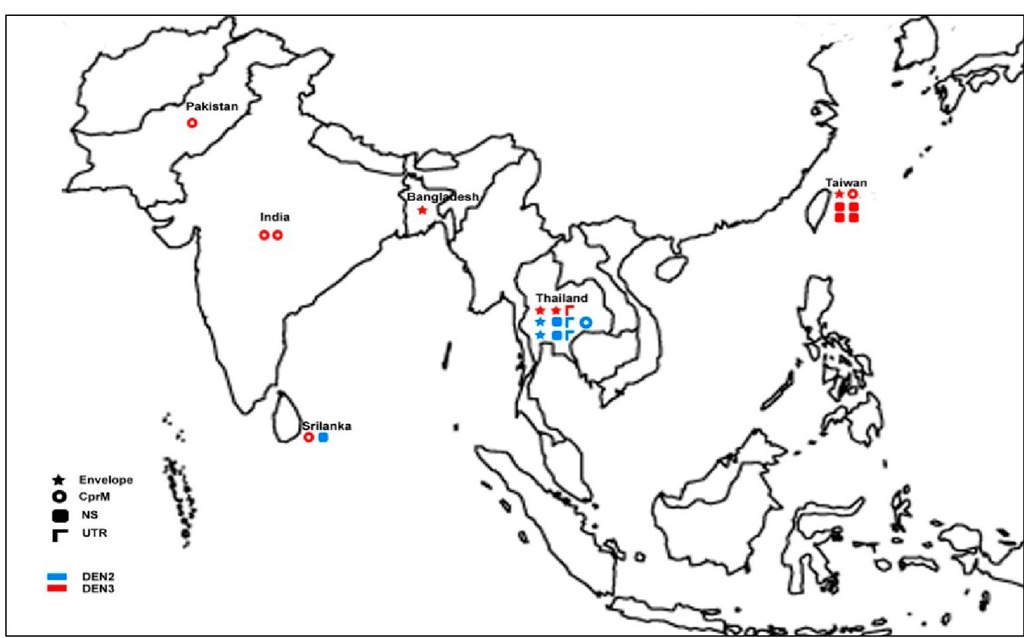
Fig. 2. The Geographical distribution of mutations in DEN-2 and DEN-3 viruses detected at different genomic loci of isolates from South East Asia
Conclusion
Ades aegypti was introduced into the coastal cities of South East Asia from East Africa around nineteenth century via the shipping industry. With the eruption of World War II it deeply entrenched in many cities. The distribution of DHF outbreaks in SEA correlates with emergence of mosquito A. egypti in South East Asian countries due to uncontrolled urbanization leading to displacement of indigenous A. albpictus from the region. Phylogenetic analysis suggests that there are foci of virus extinction and selection in South East Asian region, one such region is Thailand where the indigenous DEN-3 virus circulating up to 1992 has disappeared and replaced by two new lineages perhaps from a common ancestor. These studies point towards potential of regular extinctions of strains of dengue virus particularly DEN-3 virus and replacement by new variants in the region. Natural selection and / or genetic bottle neck are plausible causes for this variation. Since the extinction of pre 1992 strains and appearance of new epidemic strain in Thailand occurred during inter-epidemic period we therefore hypothesize that the genetic bottleneck is perhaps major cause of regional replacement. This is further supported by studies from India reporting shifting and dominance of the dengue virus serotype-3 (subtype III) replacing the earlier circulating serotype-2 (subtype IV) with emergence of increased incidence of DHF and DSS in subsequent outbreaks. Strains from the 2005 outbreak in Karachi (Pakistan) were found to be similar to those from Indian strains of dengue serotype 3, and were responsible for deadly outbreak in 2005-06.
Despite the growing genomic data base in the gene bank there are fundamental gaps in our understanding of epidemiological and evolutionary dynamics and its relation with disease severity. There are two possibilities that explain the association between clade replacement and increased viral virulence. The first is the possibility of these viruses to be better fit and therefore produce high viremia in infected humans, consequently with better transmission of virus by the vector. The other hypothesis to explain the possible virulence of emerging clades in the region is its improved ability to avoid neutralization by serotypes cross reactive antibodies (Kochel et al., 2005). Thus there is relative abundance of different serotypes and viral linage is continually changing in South East Asia. In face changing threshold of host immunity, periodic epidemics of DHF and DSS is due to local extinction and emergence of new clades. Over the period 1989 and 2000, a new genotype of DENV-1 and new clades of DENV-3 genotype III viruses have replaced older genotype and clades in this region and emergence of new clades coincided with severe epidemics.
Thus South East Asia displays greatest degree of genetic diversity, suggesting that it is the hub for the evolution of new epidemic strain. However, selection of specific clade and association of specific sequence variation with disease severity at various genomic levels reported in the literature reviewed in this study lacks strength of association i.e. reporting Relative Risk (RR)/ Odds Ratio (OR) limits our interpretation regarding causality or pin pointing specific clade with virus virulence, and therefore further studies are recommended.