Abstract
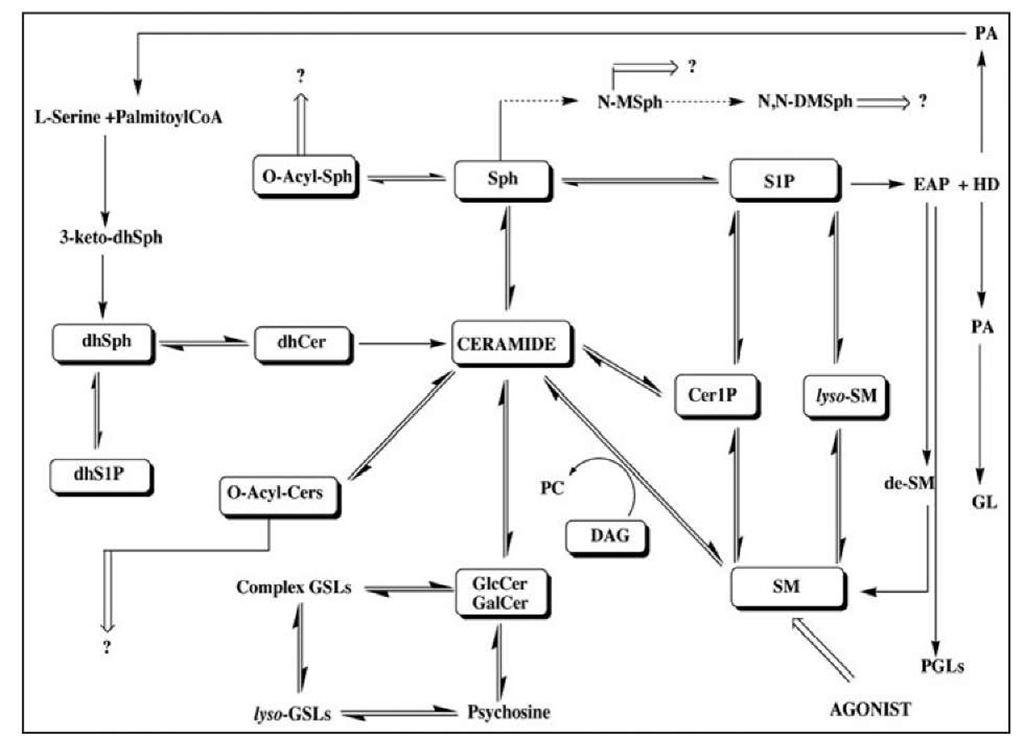
Sphingolipid (SPL) metabolism (Fig. 1) serves a key role in the complex mechanisms regulating cellular stress responses to environment. Several SPL metabolites, especially ceramide (Cer), sphingosine (Sph) and sphingosine1-phosphate (S1P) act as key bioactive molecules governing cell growth and programmed cell death (Fig. 2). Perturbations in sphingolipids of one type may enhance or interfere with the action of another. To monitor changes in SPL composition therefore, reliable analytical methods are necessary.
Here we present the liquid chromatography tandem mass spectrometry (LC-MS/MS) approach for simultaneous qualitative and quantitative monitoring of SPL components (classes and molecular species) in biological material as an effective tool to study sphingolipid signaling events. The LC-MS/MS methodology is the only available technique that provides high specificity and sensitivity, along with a wealth of structural identification information.
Introduction
Although sphingolipids (SPLs) have long been thought to function exclusively as structural constituents of the plasma membranes, in the past two decades research into the sphingolipids has progressed along two areas. First, SPLs have been shown to influence membrane structure, where they have been proposed to exist in clusters and form microdomains containing cholesterol at the plasma membrane, the so-called "lipid rafts".1 These lipid microdomains are thought to function as platforms for effective signal transduction and correct protein sorting. Second, many SPLs have been shown to act as both first and second messengers, as well as bioactive mediators, in a variety ofsignaling pathways. Thus, the SPL metabolites—ceramide (Cer), ceramide-1-phosphate (Cer1P), sphingosine (Sph) and sphingosine 1-phosphate (S1P)—have emerged as a new class of lipid biomodulators for various cell functions and through participation in and influencing of multiple signaling pathways.2-5 During the last 25 years there has been a dramatic increase in the studying of sphingolipid signaling in many patho-biological disorders but only recently new tools and approaches became available to examine these processes out, such as highly sensitive mass spectrometry methods for sphingolipid analysis. This topic describes the high-performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) for sphingolipid analysis in biological samples.
Figure 1. Sphingolipid biosynthesis and metabolic pathways; metabolomic profiling of sphingolipids. Abbreviations used in the figure: 3-keto-dhSph, 3-keto-dihydrosphingosine; dhSph, dihydrosphingosine; dhS1P, dihydrosphingosine 1-phosphate; dhCer, dihydroceramide; Sph, sphingosine; S1P, sphingosine 1-phosphate; Cer1P, ceramide-1-phosphate; SM, sphingomyelin; lyso-SM, lyso-sphingomyelin; DAG, diacylglycerol; O-Acyl-Sph. O-acyl-sphingosine, O-Acyl-Cer, O-acyl-ceramide; N-Me-Sph, N-methyl-sphingosine; N.N-DMSph, N,N-dimethyl-sphingosine; PA, palmitic acid; EAP, ethanolamine phosphate; HD, hexadecenal; GL, glycerolipids; de-SM, demethylated sphingomyelin; PGLs, phosphoglycerolipids; GlcCer, glucosylceramide; GalCer, galactosylceramide; GSLs, glycosphingolipids.
Sphingolipids: Structure and Composition
Prevalent complex SPLs: phosphosphingolipids (PSLs) and glycosphingolipids (GSLs) are found in all eukaryotes, some prokaryotes and viruses, mainly as components of the plasma membrane and related organelles. SPLs constitute about 30% of the total lipid of plasma membranes.
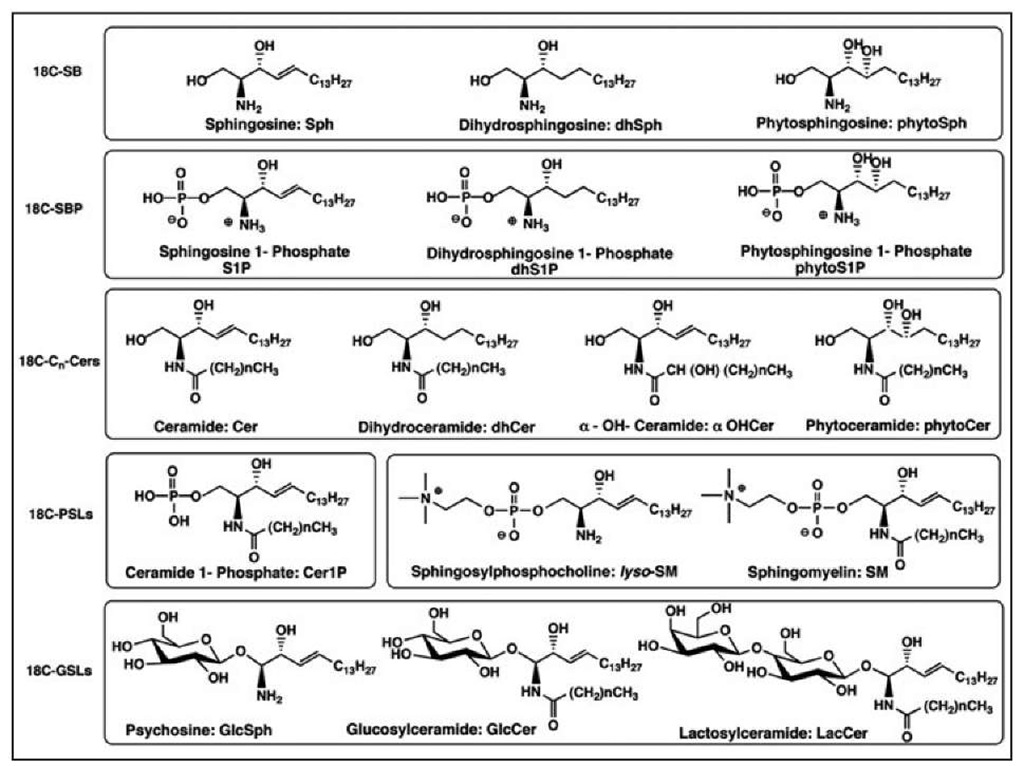
SPLs constitute one of the most structurally diversified classes of amphipathic lipids abundant in all living organisms. Variations in the nature of the head group attached to the primary hydroxyl group (carbohydrates, phosphocholine, phosphate or phosphoinositol), N-acyl group and sphingoid base (SB) backbone result in a great number of chemically distinct SPLs, where Sph, sphinganine (dhSph) or phytosphingosine (phytoSph) are the core structural moieties. Thousands of natural complex SPLs have been isolated based on almost 60 distinct species of sphingoid bases, although most of them are very minor components. SBs, the backbone of all SPLs, encompass a wide array of (2S, 3R, 4E)-2-amino-1,3-dihydroxyalkenes (Sphs), (2S, 3R)-2-amino-1,3-dihydroxy-alkanes (dhSphs) and (2S, 3S, 4R)-2-amino-1,3,4-trihydroxyalkanes (phytoSphs) with alkyl chain lengths from 14 to 22 carbon atoms and variations in the number and position of the double bonds, hydroxyl groups and branching methyl groups.
Figure 2. Natural sphingolipids are a highly heterogenous system related to the sphingoid bases and derivatization made on the amino- and hydroxy-functions. Structures shown in this figure represent derivatives of 18C-SB (sphingoid bases containing C18-backbone chain) indicating SPLs containing 2-amino-1,3-dihydroxy-octadecene-4E, 2-amino-1,3-dihydroxy-octadecane and 2-amino-1,3,4-tri-hydroxy-octadecane. General structures, nomenclature and abbreviations for SPLs are cited and described in this presentation. Cn -indicates the chain length of N-acyl part of SPLs.
Mammalian SPLs are predominantly composed of 2-amino-1,3-dihydroxyoctadecene (18CSph, abbreviated here as Sph) and 2-amino-1,3-dihydroxyoctadecane (18CdhSph, abbreviated here as dhSph) (Fig. 2). Yeast and plant SBs are mainly composed of2-amino-1,3,4-trihydroxyoctadecane (18CphytoSph), 18CdhSph and their eicosa-homologs (20CphytoSph and 20CdhSph). Additionally, some SPLs’ SBs may contain a double bond in position 8 or have double bonds in positions 4 and 8 or/and have a methyl group in position 9 of the sphingosine backbone (which can be found in plant and fungi SPLs).
Ceramides are N-acyl-derivatives of SBs. Combinations of different SBs with different fatty acids (including their hydroxy-analogs) generates a huge variety of Cers, dhCers and phytoCers.
These basic SPLs are modified at the 1-hydroxyl group to: (i) phosphates (e.g., S1P and Cer1P), (ii) phosphocholine-analogs (e.g., sphingomyelin, SM and lysosphingomyelin, lyso-SM) and (iii) glucosyl- and galactosyl-analogs (e.g., glucosylceramide, GlcCer and galactosylceramide, GalCer, known as cerebrosides and their lyso-form: psychosine). Members of the latter group also serve as precursors to hundreds of different species of complex GSPLs.
SPLs constitute the second major category of polar lipids and for example they represent approximately 5-10% of total lipid mass in mammalian brain (6-8). Abnormal SPL metabolism could lead to their accumulation and deposition in multiple tissues, especially neural tissues, that results in potentially severe clinical manifestations, known as the sphingolipidoses.6
The structural diversity of SPLs dictates that every step in analysis of these natural products must be carefully evaluated.
LC-MS Methods for Detection and Analysis of Bioactive Sphingolipids
Technological advances in lipid detection, analysis and quantitation have played a key role in promoting the development of the sphingolipid research field. Traditional lipid analytical methods, such as thin-layer chromatography, are hampered by limited sensitivity, selectivity and resolution. Metabolic labeling using lipid precursors (such as serine or palmitic acid) have been wildly used for selective labeling of certain classes of lipids, which are then typically separated using thin-layer chromatography and visualized by autoradiography. However, this method is affected by incorporation of radioactive substrates and this does not always reflect the primary lipid contents in cells. Furthermore, thin-layer chromatography has low resolution and low sensitivity; thus it is difficult to identify the subspecies of individual SPLs.
The development of advanced mass-spectrometry-based methodologies has allowed the simultaneous assessment ofseveral SPL subgroups as well as the probing of individual molecular subspecies such as various chain-length Cers. To understand the physiological function of sphingolipid metabolites, it has become important to know the metabolic change of particular SPLs and their individual subspecies from one sample. Here, we review methods for simultaneous analysis ofSPLs using liquid chromatography tandem mass spectrometry techniques.
Lipidomic Approach
The term "lipidomics" has recently emerged9-13 in relation to genomics and proteomics. Thus, lipidomics can be defined as the full characterization of lipid molecules in the studied biological material "Sphingolipidomics" will define the field of sphingolipids.
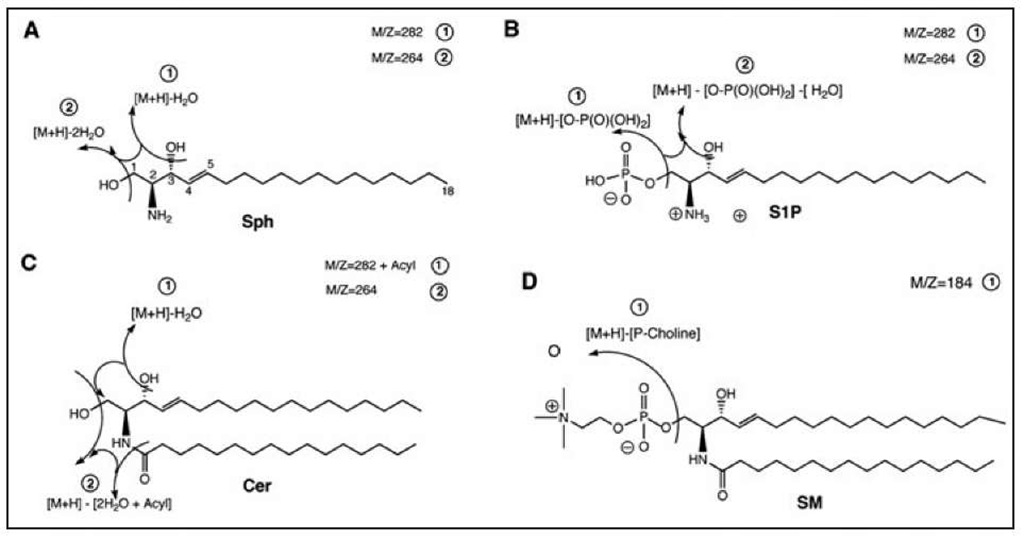
A variety of sample preparation, ionization modes and instrumental designs have been developed to analyze particular SPL classes by MS technology.14 Design for this methodology has been based on the fact that different SPL subclasses dissociate into structurally distinctive patterns corresponding to their sphingoid bases, N-acyl chains and polar headgroups.15-25 Recent advances in electrospray ionization (ESI) have provided a new approach to successfully examine total SPL components in crude lipid extracts.19,22,23,25 Electrospray ionization (ESI) methodology allows generation of intact molecular ions of molecules from solution, delivered by direct infusion or by coupling high performance liquid chromatography (HPLC) column directly to the mass spectrometer. Further improvements in instrumentation, such as the triple quadruple with robust ion sources, fast scanning mass analyzers and reduced chemical noise (mainly in MS/MS technique) allow the identification and quantitation of SPLs with great sensitivity (sub-picomol detection limit) in a highly reproducible manner. SPL identification is accomplished by tandem mass spectrometry (MS/MS) with precursor ion scans to distinguish various molecular species in crude lipid extract by taking advantage of the unique molecular decomposition pattern19,25 for each SPL class (Fig. 3). SPL quantitation is performed by using positive ionization and multiple reaction monitoring (MRM) in conjunction with HPLC separation.25 Liquid chromatography/ tandem mass spectrometry (LC-MS/MS) is the only technology available that provides structural specificity, quantitative precision and relatively high-throughput for analysis of complex SPLs in small samples.
Sample Preparation
The extraction process is one of the most important steps in pretreatment of solid (cell pellets, tissue) and liquid (plasma, serum, whole blood, biological fluids) samples.
Chloroform: methanol 2:1 (v/v) extraction, developed in 1956 for fish tissue,26 further improved in 1959 by Bligh and Dyer,27 became a golden standard procedure, known as the "Bligh & Dyer" (B & D) method and it is still commonly used for lipid extraction from all biological matrices. It involves a two step extraction employing chloroform: methanol: water at well-defined ratios of 1:2:0.8 and 2:2:1.8, respectively. According to the originators, the upper (methanol: water) phase contains all the "nonlipid" substances, while most lipids remain in the lower (chloroform) phase. This virtually unchanged procedure is commonly applied to most SPL sample preparation, regardless of the analytical procedure subsequently used e.g., TLC, HPLC or MS, although the extraction efficiency, particularly for the most polar SPL components as S1P or lyso-SM seems to be questionable.
Figure 3. Typical MS/MS fragmentation patterns of sphingosine (A), sphingosine 1-phosphate (B), ceramide (C) and sphingomyelin (D). The specific common fragment ion of m/z = 264 (2, panel C) for Cers and m/z = 184 (1, panel D) for SM are used in the Parent Ion Scan experiments for determination of molecular species composition prior to quantitative analysis with the MRM experiments.
Over time, some modifications to the B & D have been incorporated in the isolation of SPLs, mostly intended to remove the bulk of the major co-extracted components, especially the glycero-lipids, by subjecting the initial chloroform extract to a mild alkaline hydrolysis that cleaves ester linkage.25,28-36 However, 1-O-acyl-ceramides (O-AcylCer) and related compounds37 will also be hydrolyzed, thus artificially increasing the level of Cers. Comparison of the Cer level calculated from lipid extracts that were prepared with and without the base hydrolysis step can provide important data about the level of O-AcylCers. Our results showed some (20-40%) increase in Cers after this treatment. Nevertheless, this simple approach is recommended in the preparation of samples for SM analysis to allow elimination of phosphatidylcholine (PC) from the lipid extract, which may interfere with SM determination, even at highly specific LC-MS/MS analysis, due to close masses and fragmentation pattern.
Several attempts to further separate the initial total lipid extracts into particular lipid classes by a set of solid phase extraction (SPE) cartridges38-40 proved to be very time consuming and not reproducible. Moreover, it may not be necessary when selective LC/MS technology is employed for analysis.
We developed41 a one-phase extraction, using ethyl acetate: iso-propanol: water system at 60:30:10; (v/v/v) and 85:15:0 (v/v/v) for cell pellets and tissue homogenates and aqueous samples, respectively. The protocol describes lipid extraction under a safe and neutral condition to avoid destruction of the parent "soft" SPLs (e.g., SPLs containing O-acyl group), assuring efficient and quantitative extraction of the SB-1Ps from biological material since the latter are notoriously difficult to recover quantitatively.39,42
Readers interested in developing and/or improving existing methods of sample preparation are referred to an excellent review by McDowall.43