The HLA Complex and Its Products
The human major histocompatibility complex (MHC), commonly called the human leukocyte antigen (HLA) complex, is a 4-megabase (Mb) region on chromosome 6 (6p21.3) that is densely packed with expressed genes. The best known of these genes are the HLA class I and class II genes, whose products are critical for immunologic specificity and transplantation histocompatibility, and they play a major role in susceptibility to a number of autoimmune diseases. Many other genes in the HLA region are also essential to the innate and antigen-specific functioning of the immune system. The HLA region shows extensive conservation with the MHC of other mammals in terms of genomic organization, gene sequence, and protein structure and function. Much of our understanding of the MHC has come from investigation of the MHC in mice, which is termed the H-2 complex, and to a lesser degree from other species as well. Nonetheless, in this topic discussion is confined to information applicable to the MHC in humans.
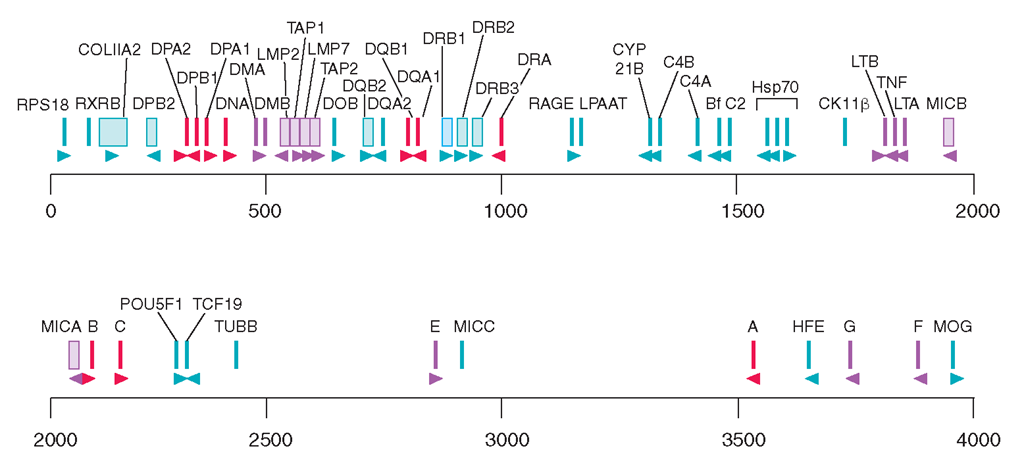
The HLA class I genes are located in a 2-Mb stretch of DNA at the telomeric end of the HLA region (Fig. 2-1). The classic (MHC class Ia) HLA-A, -B, and -C loci, the products of which are integral participants in the immune response to intracellular infections, tumors, and allografts, are expressed in all nucleated cells and are highly polymorphic in the population. Polymorphism refers to a high degree of allelic variation within a genetic locus that leads to extensive variation between different individuals expressing different alleles. Over 450 alleles at HLA-A, 780 at HLA-B, and 230 at HLA-C have been identified in different human populations, making this the most highly polymorphic segment known within the human genome. Each of the alleles at these loci encodes a heavy chain (also called an α chain) that associates noncovalently with the nonpolymorphic light chain ß2-microglobulin, encoded on chromosome 15.
The nomenclature of HLA genes and their products reflects the grafting of newer DNA sequence information on an older system based on serology. Among class I genes, alleles of the HLA-A, -B, and -C loci were originally identified in the 1950s, 1960s, and 1970s by alloantisera, derived primarily from multiparous women, who in the course of normal pregnancy produce antibodies against paternal antigens expressed on fetal cells. The serologic allotypes were designated by consecutive numbers, e.g., HLA-A1, HLA-B8. Currently, under World Health Organization (WHO) nomenclature, class I alleles are given a single designation that indicates locus, serologic specificity, and sequence-based subtype. For example, HLA-A*0201 indicates subtype 1 of the serologically defined allele HLA-A2. Subtypes that differ from each other at the nucleotide but not the amino acid sequence level are designated by an extra numeral, e.g., HLA-B*07021 and HLA-B*07022 are two variants of the HLA-B702 subtype of HLA-B*07.
FIGURE 2-1
Physical map of the HLA region, showing the class I and class II loci, other immunologically important loci, and a sampling of other genes mapped to this region. Gene orientation is indicated by arrowheads. Scale is in kilobase (kb).
The nomenclature of class II genes, discussed below, is made more complicated by the fact that both chains of a class II molecule are encoded by closely linked HLA-encoded loci, both of which may be polymorphic, and by the presence of differing numbers of isotypic DRB loci in different individuals. It has become clear that accurate HLA genotyping requires DNA sequence analysis, and the identification of alleles at the DNA sequence level has contributed greatly to the understanding of the role of HLA molecules as peptide-binding ligands, to the analysis of associations of HLA
The approximate genetic distance from DP to A is 3.2 cM. This includes 0.8 cM between A and B (including 0.2 cM between C and B), 0.4-0.8 cM between B and DR-DQ, and 1.6-2.0 cM between DR-DQ and DP. alleles with certain diseases, to the study of the population genetics of HLA, and to a clearer understanding of the contribution of HLA differences to allograft rejection and graft-vs-host disease. Current databases of HLA class I and class II sequences can be accessed by Internet (e.g., from the IMGT/HLA Database, http://uuu.ebi.ac.uk/ imgt/hla), and frequent updates of HLA gene lists are published in several journals.
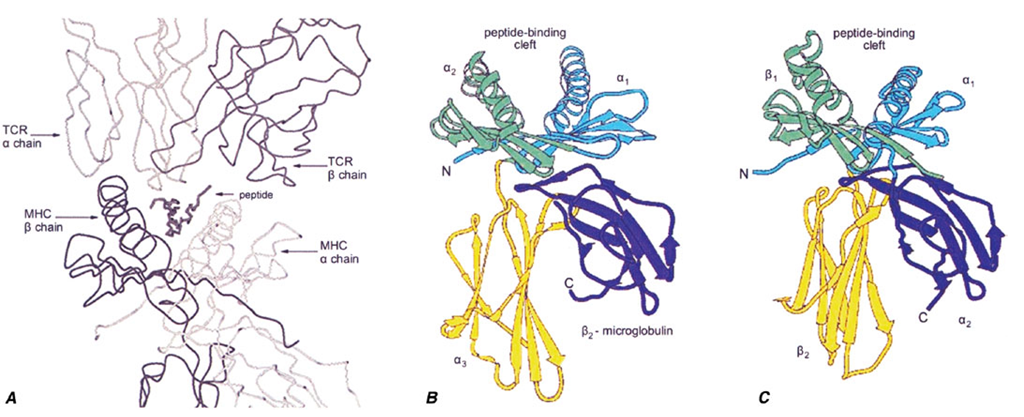
The biologic significance of this MHC genetic diversity, resulting in extreme variation in the human population, is evident from the perspective of the structure of MHC molecules.As shown in Fig. 2-2, the MHC class I and class II genes encode MHC molecules that bind small peptides, and together this complex (pMHC; peptide-MHC) forms the ligand for recognition by T lymphocytes, through the antigen-specific T cell receptor. There is a direct link between the genetic variation and this structural interaction: The allelic changes in genetic sequence result in diversification of the peptide-binding capabilities of each MHC molecule and in differences for specific TCR binding. Thus, different pMHC complexes bind different antigens and are targets for recognition by different T cells.
FIGURE 2-2
A. The trimolecular complex of TCR (top), MHC molecule (bottom), and a bound peptide form the structural determinants of specific antigen recognition. Other panels (B and C) show the domain structure of MHC class I (B) and class II (C) molecules. The α! and α2 domains of class I and the α! and ßi domains of class II form a ß-sheet platform that forms the floor of the peptide-binding groove, and α helices that form the sides of the groove. The α3 (A) and ß2 domains (B) project from the cell surface and form the contact sites for CD8 and CD4, respectively.
The class I MHC and class II MHC structures, shown in Figs. 2-2B and 2-2C, are structurally closely related, although there are a few key differences. While both bind peptides and present them to T cells, the binding pockets have different shapes, which influences the types of immune responses that result (discussed below). In addition, there are structural contact sites for T cell molecules known as CD8 and CD4, expressed on the class I or class II membrane-proximal domains, respectively. This ensures that when peptide antigens are presented by class I molecules, the responding T cells are predominantly of the CD8 class, and similarly, that T cells responding to class II pMHC complexes are predominantly CD4.
The nonclassic, or class Ib, MHC molecules, HLA-E, -F, and -G, are much less polymorphic than MHC Ia and appear to have distinct functions. The HLA-E molecule, which has a peptide repertoire restricted to signal peptides cleaved from classic MHC class I molecules, is the major self-recognition target for the natural killer (NK) cell inhibitory receptors NKG2A or NKG2C paired with CD94 (see below and Chap. 1); four HLA-E alleles are known. HLA-G is expressed selectively in extravillous trophoblasts, the fetal cell population directly in contact with maternal tissues. It binds a wide array of peptides, is expressed in six different alternatively spliced forms, and provides inhibitory signals to both NK cells and T cells, presumably in the service of maintaining maternofetal tolerance. The function of HLA-F remains largely unknown.
Additional class I—like genes have been identified, some HLA-linked and some encoded on other chromosomes, that show only distant homology to the class Ia and Ib molecules but share the three-dimensional class I structure. Those on chromosome 6p21 include MIC-A and MIC-B, which are encoded centromeric to HLA-B, and HLA-HFE, located 3 to 4 cM (centi-Morgan) telomeric of HLA-F. MIC-A and MIC-B do not bind peptide but are expressed on gut and other epithelium in a stress-inducible manner and serve as activation signals for certain γδ T cells, NK cells, CD8 T cells, and activated macrophages, acting through the activating NKG2D receptors. Sixty-one MIC-A and twenty-five MIC-B alleles are known, and additional diversification comes from variable alanine repeat sequences in the transmembrane domain. HLA-HFE encodes the gene defective in hereditary hemochromatosis. Among the non-HLA, class I—like genes, CD1 refers to a family of molecules that present glycolipids or other nonpeptide ligands to certain T cells, including T cells with NK activity; FcRn binds IgG within lysosomes and protects it from catabolism (Chap. 1); and Zn-O2-Blycoprotein 1 binds a nonpeptide ligand and promotes catabolism of triglycerides in adipose tissue. Like the HLA-A, -B, -C, -E, -F, and -G heavy chains, each of which forms a heterodimer with ß2-microglobulin (Fig. 2-2), the class I—like molecules, HLA-HFE, FcRn, and CD1 also bind to ß2-microglobulin, but MIC-A, MIC-B, and Zn-O2-glycoprotein 1 do not.
The HLA class II region is also illustrated in Fig. 2-1. Multiple class II genes are arrayed within the centromeric 1 Mb of the HLA region, forming distinct haplotypes. A haplotype refers to an array of alleles at polymorphic loci along a chromosomal segment. Multiple class II genes are present on a single haplotype, clustered into three major subregions: HLA-DR, -DQ, and -DP Each of these subregions contains at least one functional alpha (A) locus and one functional beta (B) locus. Together these encode proteins that form the α and β polypeptide chains of a mature class II HLA molecule. Thus, the DRA and DRB genes encode an HLA-DR molecule; products of the DQAl and DQBl genes form an HLA-DQ molecule; and the DPAl and DPBl genes encode an HLA-DP molecule. There are several DRB genes (DRB1, DRB2, DRB3, etc.), so that two expressed DR molecules are encoded on most haplotypes by combining the α-chain product of the DRA gene with separate β chains. More than 438 alleles have been identified at the HLA-DRB1 locus, with most of the variation occurring within limited segments encoding residues that interact with antigens. Detailed analysis of sequences and population distribution of these alleles strongly suggests that this diversity is actively selected by environmental pressures associated with pathogen diversity.
The class II region was originally termed the D-region. The allelic gene products were first detected by their ability to stimulate lymphocyte proliferation by mixed lymphocyte reaction and were named Dw1, Dw2, etc. Subsequently, serology was used to identify gene products on peripheral blood B cells, and the antigens were termed DR (D-related). After additional class II loci were identified, these came to be known as DQ and DP. In the DQ region, both DQA1 and DQB1 are polymorphic, with 34 DQA1 alleles and 71 DQB1 alleles. The current nomenclature is largely analogous to that discussed above for class I, using the convention “locus*allele.” Thus, for example, subtypes of the serologically defined specificity DR4, encoded by the DRB1 locus, are termed DRB1*0401, -0402, etc. In addition to allelic polymorphism, products of different DQA1 alleles can, with some limitations, pair with products of different DQB1 alleles through both cis and trans pairing to create combinatorial complexity and expand the number of expressed class II molecules. Because of the enormous allelic diversity in the general population, most individuals are heterozygous at all of the class I and class II loci. Thus, most individuals express six classic class I molecules (two each of HLA-A, -B, and -C) and around eight class II molecules—two DP, two DR (more in the case of haplotypes with additional functional DRB genes), and up to four DQ (two cis and two trans).
Other Genes in the MHC
In addition to the class I and class II genes themselves, there are numerous genes interspersed among the HLA loci that have interesting and important immunologic functions. Our current concept of the function of MHC genes now encompasses many of these additional genes, some of which are also highly polymorphic. Indeed, direct comparison of the complete DNA sequences for two of the entire 4-Mb MHC regions from different haplotypes show >18,000 variations, encoding an extremely high potential for biologic diversity. Specific examples include the TAP and LMP genes, as discussed in more detail below, which encode molecules that participate in intermediate steps in the HLA class I biosynthetic pathway. Another set of HLA genes, DMA and DMB, perform an analogous function for the class II pathway. These genes encode an intracellular molecule that facilitates the proper complexing of HLA class II molecules with antigen (see below). The HLA class III region is a name given to a cluster of genes between the class I and class II complexes, which includes genes for the two closely related cytokines tumor necrosis factor (TNF)^ and lymphotoxin (TNF-ß); the complement components C2, C4, and Bf; heat shock protein (HSP)70; and the enzyme 21-hydroxylase.
The class I genes HLA-A, -B, and -C are expressed in all nucleated cells, although generally to a higher degree on leukocytes than on nonleukocytes. In contrast, the class II genes show a more restricted distribution: HLA-DR and HLA-DP genes are constitutively expressed on most cells of the myeloid cell lineage, whereas all three class II gene families (HLA-DR, -DQ, and -DP) are inducible by certain stimuli provided by inflammatory cytokines such as interferon γ. Within the lymphoid lineage, expression of these class II genes is constitutive on B cells and inducible on human T cells. Most endothelial and epithelial cells in the body, including the vascular endothelium and the intestinal epithelium, are also inducible for class II gene expression. Thus, while these somatic tissues normally express only class I and not class II genes, during times of local inflammation they are recruited by cytokine stimuli to express class II genes as well, thereby becoming active participants in ongoing immune responses. Class II expression is controlled largely at the transcriptional level through a conserved set of promoter elements that interact with a protein known as CIITA. Cytokine-mediated induction of CIITA is a principal method by which tissue-specific expression of HLA gene expression is controlled. Other HLA genes involved in the immune response, such as TAP and LMP, are also susceptible to upregulation by signals such as interferon γ. Sequence data for the entire HLA region can be accessed on the Internet (e.g., http://uuu.sanger.ac.uk/HGP/Chr6/MHC). Many new genes have been discovered, the functions of which remain to be determined, as well as numerous microsatellite regions and other genetic elements. The gene density of the class II region is high, with approximately one protein encoded every 30 kb, and that of the class I and class III regions is even higher, with approximately one protein encoded every 15 kb.
Linkage Disequilibrium
In addition to extensive polymorphism at the class I and class II loci, another characteristic feature of the HLA complex is linkage disequilibrium. This is formally defined as a deviation from Hardy-Weinberg equilibrium for alleles at linked loci. This is reflected in the very low recombination rates between certain loci within the HLA complex. For example, recombination between DR and DQ loci is almost never observed in family studies, and characteristic haplotypes with particular arrays of DR and DQ alleles are found in every population. Similarly, the complement components C2, C4, and Bf are almost invariably inherited together, and the alleles at these loci are found in characteristic haplo-types. In contrast, there is a recombinational hotspot between DQ and DP, which are separated by 1-2 cM of genetic distance, despite their close physical proximity. Certain extended haplotypes encompassing the interval from DQ into the class I region are commonly found, the most notable being the haplotype DR3-B8-A1, which is found, in whole or in part, in 10-30% of northern European Caucasians. It has been hypothesized that selective pressures may maintain linkage disequilibrium in HLA, but this remains to be determined. As discussed below under HLA and immunologic disease, one consequence of the phenomenon of linkage disequilibrium has been the resulting difficulty in assigning HLA-disease associations to a single allele at a single locus.
MHC Structure and Function
Class I and class II molecules display a distinctive structural architecture, which contains specialized functional domains responsible for the unique genetic and immunologic properties of the HLA complex.The principal known function of both class I and class II HLA molecules is to bind antigenic peptides in order to present antigen to an appropriate T cell. The ability of a particular peptide to satisfactorily bind to an individual HLA molecule is a direct function of the molecular fit between the amino acid residues on the peptide with respect to the amino acid residues of the HLA molecule. The bound peptide forms a tertiary structure called the MHC-peptide complex, which communicates with T lymphocytes through binding to the T cell receptor (TCR) molecule. The first site of TCR-MHC-peptide interaction in the life of a T cell occurs in the thymus, where self-peptides are presented to developing thymocytes by MHC molecules expressed on thymic epithelium and hematopoietically derived antigen-presenting cells, which are primarily responsible for positive and negative selection, respectively (Chap. 1). Thus, the population of MHC-T cell complexes expressed in the thymus shapes the TCR repertoire. Mature T cells encounter MHC molecules in the periphery both in the maintenance of tolerance (Chap. 3) and in the initiation of immune responses. The MHC-peptide-TCR interaction is the central event in the initiation of most antigen-specific immune responses, since it is the structural determinant of the specificity. For potentially immunogenetic peptides, the ability of a given peptide to be generated and bound by an HLA molecule is a primary feature of whether or not an immune response to that peptide can be generated, and the repertoire of peptides that a particular individual’s HLA molecules can bind exerts a major influence over the specificity of that individual’s immune response.
When a TCR molecule binds to an HLA-peptide complex, it forms intermolecular contacts with both the antigenic peptide and with the HLA molecule itself. The outcome of this recognition event depends on the density and duration of the binding interaction, accounting for a dual specificity requirement for activation of the T cell.That is, the TCR must be specific both for the antigenic peptide and for the HLA molecule. The polymorphic nature of the presenting molecules, and the influence that this exerts on the peptide repertoire of each molecule, results in the phenomenon of MHC restriction of the T cell specificity for a given peptide. The binding of CD8 or CD4 molecules to the class I or class II molecule, respectively, also contributes to the interaction between the T cell and the HLA-peptide complex, by providing for the selective activation of the appropriate T cell.
Class I Structure
As noted above, MHC class I molecules provide a cell-surface display of peptides derived from intracellular proteins, and they also provide the signal for self-recognition by NK cells. Surface-expressed class I molecules consist of an MHC-encoded 44-kD glycoprotein heavy chain, a non-MHC-encoded 12-kD light chain ß2-microglobulin, and an antigenic peptide, typically 8-11 amino acids in length and derived from intracellularly produced protein. The heavy chain displays a prominent peptide-binding groove. In HLA-A and -B molecules, the groove is ~3 nm in length by 1.2 nm in maximum width (30 A X 12 A), whereas it is apparently somewhat wider in HLA-C. Antigenic peptides are noncovalently bound in an extended conformation within the peptide-binding groove, with both N- and C-terminal ends anchored in pockets within the groove (A and F pockets, respectively) and, in many cases, with a prominent kink, or arch, approximately one-third of the way from the N-terminus that elevates the peptide main chain off the floor of the groove.
A remarkable property of peptide binding by MHC molecules is the ability to form highly stable complexes with a wide array of peptide sequences. This is accomplished by a combination of peptide sequence-independent and peptide sequence-dependent bonding. The former consists of hydrogen bond and van der Waals interactions between conserved residues in the peptide-binding groove and charged or polar atoms along the peptide backbone. The latter is dependent upon the six side pockets that are formed by the irregular surface produced by protrusion of amino acid side chains from within the binding groove. The side chains lining the pockets interact with some of the peptide side chains. The sequence polymorphism among different class I alleles and isotypes predominantly affects the residues that line these pockets, and the interactions of these residues with peptide residues constitute the sequence-dependent bonding that confers a particular sequence “motif” on the range of peptides that can bind any given MHC molecule.