Protein interactions occur in many different forms: Proteins interact with and modify each other as elements of signaling chains; they form large molecular machines with more than 100 protein and nonprotein elements, for example, the spliceosome; or, they assemble to provide structural elements of the cell. Proteins interact not only with each other but also with DNA, RNA, and other molecule types. Types of interaction may vary from direct physical contact to very indirect interactions, for example, participation in the same signaling pathway. In many contexts, protein states, in particular, posttranslational modifications, are essential for the function of protein assemblies.
The kind of data produced by experimental technologies to elucidate protein interaction data is similarly diverse, from direct, atom-level structural information by NMR (Pellecchia et al., 2002) or X ray via evidence for direct physical contact, for example, by yeast two hybrid assays (Phizicky et al., 2003) and information on participation in the same complex by tandem affinity purification (Rigaut et al., 1999) to indirect information, for example, colocalization.
With the introduction of technologies for high-throughput interaction screens (Uetz et al., 2000; Ito et al., 2001; Gavin et al., 2002; Ho et al., 2002), the number of interactions published in a single publication now varies over four orders of magnitude, from single interactions to more than 20 000 (Giot et al., 2003).
The biological and medical importance of protein interactions, high-throughput technologies, and the broadness of data types and experimental technologies have led to the creation of a significant number of protein interaction data collections, from simple HTML tables providing the results of a particular experiment to large databases providing complex tools. We subsequently introduce some criteria for the comparison of protein interaction databases, and describe some major, publicly accessible protein interaction databases on the basis of these criteria (Table 1), focusing on databases for experimentally, not computationally, derived datasets.
Confidence information: High-throughput experiments are often considered less reliable than detailed, low-throughput experiments. To support the user in estimating the reliability of individual interactions, the authors of large-scale interaction data sets often provide quality indications, for example, the grouping into reliability classes (Li et al., 2004) or numerical values (Giot et al., 2003). In addition, comparative analysis of different interaction data sets provides quality indications. In the simplest case, this is the number of times an interaction has been observed in independent experiments, but quality assessments are also based, for example, on the comparison against a reference data set considered to be correct (von Mering et al., 2002), shared subcellular location and cellular role annotation of interacting proteins (Sprinzak et al., 2003), comparison to RNA expression profiles, or similar interactions in paralogous sequences (Deane et al., 2002).
Table 1 Contents and features of publicly available protein interaction databases
| Acronym | BIND | CYGD |
| Name | Biomolecular Interaction Network Database | Comprehensive Yeast Genome Database |
| Description | The Biomolecular Interaction Network Database (BIND) is a collection of records documenting molecular interactions. The contents of BIND include high-throughput data submissions and hand-curated information gathered from the scientific literature. | The MIPS Comprehensive Yeast Genome Database (CYGD) aims to present information on the molecular structure and functional network of the entirely sequenced, well-studied model eukaryote, the budding yeast Saccharomyces cerevisiae. |
| Contents | 94 368 interactions (including genetic interactions) | 15 488 (9103 physical and 6385 genetic) |
| Species range | All | Saccharomyces cerevisiae |
| Search | Simple search box. and
field-specific search interface for complex queries. Search by GI. gene name. PubMed id. protein description. Blast search. Returns 21 interactions for “lsm7″. |
Simple search box. Search by ORF/gene name or PubMed id. Cross-referenced from well-annotated Yeast Genome Database. |
| Visualization | Java WebStart application. Network viewer with unusual “ontoglyphs” to visualize protein properties. Support for Cytoscape in download files. | The CYGD site was being updated at the time of writing, the visualization system could not be accessed. |
| DIP | GRID |
| Database of Interacting Proteins | General Repository for Interaction Datasets |
| The DIP database catalogs experimentally determined interactions between proteins. It combines information from a variety of sources to create a single, consistent set of protein-protein interactions. | The GRID is a database of genetic and physical interactions developed in the Tyers Group at the Samuel Lunenfeld Research Institute at Mount Sinai Hospital. |
| 44 349 protein interactions | 54 000 genetic and protein interactions |
| All (107 organisms)
Four search forms. Search by description, prosite motif or publication. Blast search. |
Yeast. Fly. Worm. Human. Mouse.
Zebrafish, S. pombe, and Rat Simple search box. Search by GI number. ORF/Gene name. Search for “lsm7″ returns 20 |
| Search by “lsm7″ returned no result, recommended way is to search UniProt, then use the link there, or the sequence. | interaction partners. |
| Graph view with interaction reliability projected on edge colors. | “Osprey” application. Requires local installation, provides powerful graph exploration and integration of user-supplied data. No connection to GRID search. Osprey search only on current dataset. |
| Confidence information | Indirect, number of PubMed abstracts per interaction. | Not available. |
| Data availability | Free to academic and commercial users. | Free to academic users. License required for commercial use. No redistribution. |
| Data structure | Highly detailed, complex data structure. Based on NCBI data types. Published and available from website. | Database structure unpublished. |
| Download formats | Various full and subset files in ASN.l. Fasta. XML (BIND-specific) format | Tab-delimited files. |
| Software availability | Mainly C/C++, source code available. | Not available. |
| Documentation | Well-documented, including data submission and curation manuals. | Project descriptions. |
| URL | http://www.blueprint.org/bincl/ bind.php | http ://mips.gsf.de |
| Reference Notes | Bader et al. (2003) | Mewes et al. (2004) |
| Visited on | July 2. 2004 | July 5. 2004 |
| Interaction confidence | Indirect, list of publications. |
| classification according to three different methods. Grouping of interactions into reliable “core” and less reliable “noncore”. | |
| Free to academic end users, no redistribution allowed. Flat file download after registration. License required for commercial use. | Free to academic end users, no redistribution allowed. Flat file download after registration. License required for commercial use. |
| Relatively simple relational table structure. Overview described in Salwinski et al. (2004). | Tab-delimited format. |
| XIN (DIP-specific) XML format. PSI-MI XML. Fasta | Tab-delimited format. |
| Not available. | Not available. |
| Usage and search guide documents. | No Grid documentation, but message-based support forum. On-line manual for Osprey. |
| http://dip.doe-
mbi.ucla.edu/dip/Main.cgi |
http://biodata.mshri.on.ca/grid/ servlet/Index |
| Salwinski et al. (2004) Separate satellite database providing information on protein state. | Breitkreutz et al. (2003) |
| July 5. 2004 | July 5. 2004 |
Table 1 (continued)
| Acronym | HPRD | Hybrigenics |
| Name | Human Protein Reference Database | Hybrigenics S.A. |
| Description | The Human Protein Reference Database represents a centralized platform to visually depict and integrate | PIMRider® is Hybrigenics’ functional proteomics software platform, dedicated to the exploration of protein |
| information pertaining to domain architecture, posttranslational modifications, interaction networks, and disease association for each protein in the human proteome. | pathways. Based on reliable Protein Interaction Maps (PIM®), PIMRider® leads to the unraveling of biological functions. | |
| Contents Species range | 15 944 protein interactions Human | 4200 protein interactions Human, Drosophila, H. pylori |
| Search | Multifield search form. Search by protein name, gene name, posttranslational modification, GO, and domain annotation of proteins. | Simple search box. Search by gene name/description. |
| Visualization | Visualization of interacting domains. No graph-oriented visualization. | PIMRider® visualization tool. Java-based for local installation. Graph view, linked to view or interacting domains. Filtering on confidence score (PBS). |
| Confidence information | Indirect, by indication of experiment type. | Full integration of PBS score Rain et al. (2001) |
| IntAct | MINT |
| Open Source Database of Molecular Interactions | Molecular Interactions Database |
| IntAct provides an open source database and toolkit for the storage, presentation, and analysis of protein interactions. IntAct data is curated from large and small-scale | MINT is a relational database designed to store interactions between biological molecules. Presently. MINT focuses on experimentally verified protein interactions with special |
| experiments. | emphasis on proteomes from mammalian organisms. |
| 37 680 protein interactions All (120 species) | 42 534 interactions All |
| Simple search box. Search by gene name. UniProt, InterPro, GO, PubMed, model organism database accession numbers. | Multifield search form. Search by UniProt. InterPro, PDB, GO, PubMed identifiers, gene names, UniProt keywords. |
| Graph visualization, highlighting of graph nodes according to GO annotation. | Java Applet Mint viewer with visualization of number of times an interaction has been observed. |
| Indirect, by number of experiments. | Indirect, by number of experiments. |
| Data availability | Free to academic users. File download after registration. License required for commercial use. | TGF-Beta, Drosophila datasets free for all users after registration. H.pylori dataset free to academic users. File download after registration. License required for commercial use. |
| Data structure | Moderately complex, no detail documentation, only autogenerated UML diagram. | Not documented. |
| Download formats | HRPD-specific XML format. PSI-MI XML. | PSI-MI XML. |
| Software availability | Open source availability
announced, but not yet there. |
PIMWalker tool (PIMRider with reduced features, but capable of visualizing PSI-MI files) available for noncommercial use after registration. Contains partial source code. |
| Documentation | Extensive FAQ documents. | Compact, but comprehensive PIMRider on-line manual. |
| URL | http://www.hprd.org/ | http ://pim.hyb rigenics.com/ |
| Reference | Peri et al. (2004) | Rain et al. (2001) |
| Notes | ||
| Visited on | July 5. 2004 | July 6. 2004 |
| Free to academic and commercial users. | File download after registration. No statement on commercial use of redistribution restrictions. |
| Moderately complex. UML diagram and relational schema available on the web. | Data structure follows the PSI-MI standard. |
| PSI-MI XML. GO formatted controlled vocabularies. | PSI-MI XML. |
| Open source, well-documented and freely available, with installation instructions. | Not available. |
| Detailed user manual. | On-line documentation. |
| http//www.ebi.ac.uk/intact | http://inint.bio.uniroma2.it/iiiint/ |
| Hermjakob et al. (2004b) Dynamic download of interaction networks in PSI-MI format, for example, for Cytoscape support. | Zanzoni et al. (2002) |
| July 5. 2004 | July 6. 2004 |
Standards: The number of interactions in the yeast Saccharomyces cerevisiae interactome has been estimated to be about 10 000-26 000 (Sprinzak et al., 2003; Grigoriev, 2003). Taking into account all relevant species and the change of protein interactions depending on protein and cellular state, the number of interactions is nearly unlimited. As it is unlikely that any given database will be able to collect all available protein interaction data, collecting data from several, potentially specialized databases is essential to assemble a reasonably complete picture of the currently available protein interaction data in a given domain. In addition to providing a complete picture, such collections also provide the basis for interaction data reliability assessments through comparative analysis. However, such collection may be difficult and labor-intensive due to different data formats and annotations used by different databases. To improve this situation, major interaction data providers, among them BIND (Biomolecular Interaction Network Database), DIP (Database of Interacting Proteins), Hybrigenics, HPRD (Human Protein Reference Database), IntAct, MIPS, and MINT, in the framework of the HUPO Proteomics Standards Initiative (http://psidev.sf.net) (see Article 61, Data standardization and the HUPO proteomics standards initiative, Volume 7) have jointly developed the PSI-MI XML format, a community standard for the representation of protein interaction data. PSI-MI 1.0 provides a basic exchange format for protein interaction data (Hermjakob et al., 2004a); level 2.0 is under development (Orchard et al., 2004) and will provide additional features and an extension to additional molecule types, in particular, RNA and DNA. In addition to a standard format, PSI-MI provides a set of interaction-specific controlled vocabularies to standardize not only the format but also the contents of protein interaction data, for example, to select all data derived by a certain technology.
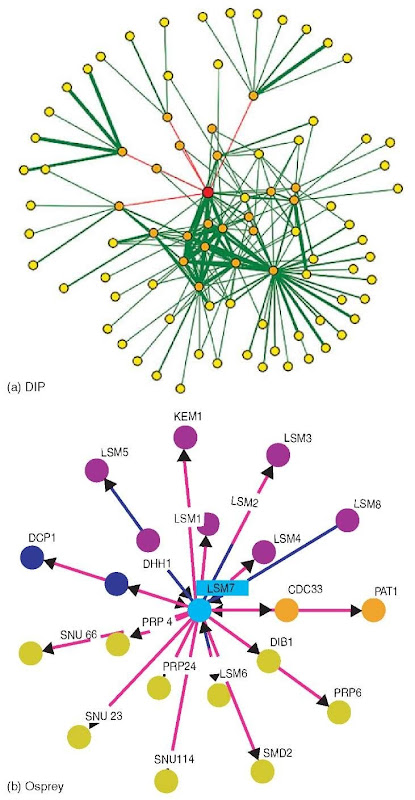
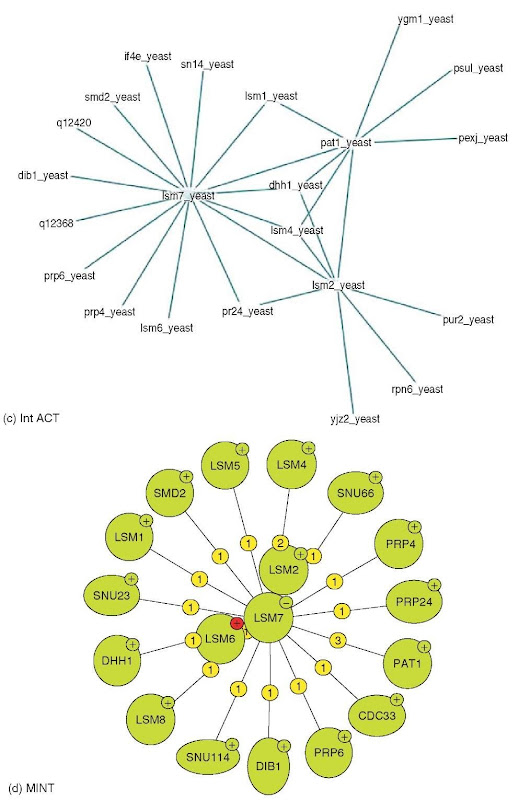
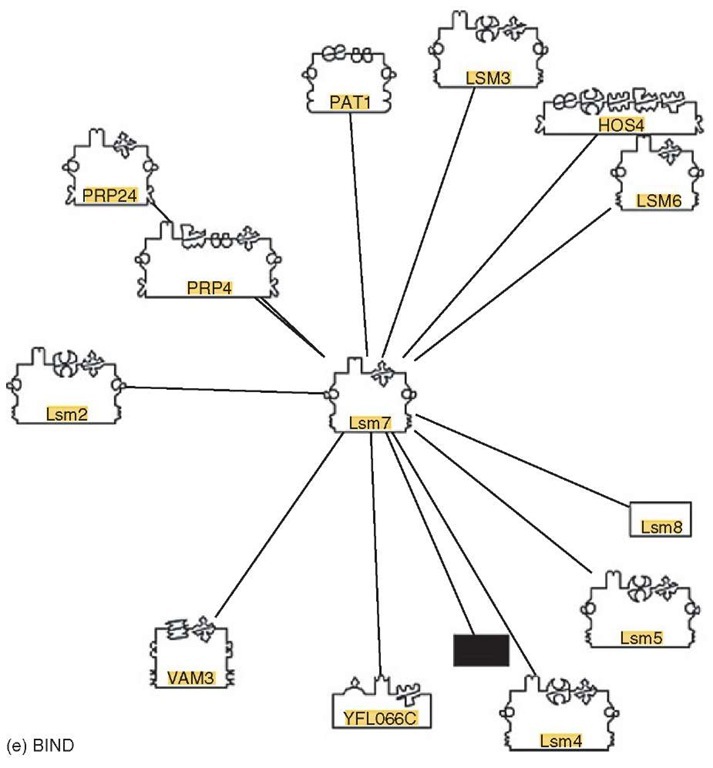
Visualization: In scientific publications, protein interactions are often visualized as groups of adjacent shapes with textual annotation. While such representations are intuitive, they are difficult to generate automatically. For automatically generated visualization, nearly all tools are based on the abstraction of proteins as nodes and interactions as edges in a graph. This representation allows the application of well-established graph layout algorithms to interaction networks, and provides the basis for additional analysis types, for example, interaction distance, or the identification of clusters of highly interconnected proteins. Although mostly based on this basic abstraction, tools differ significantly in the technical implementation, user interface, and, in particular, in methods to project additional information onto such interaction networks, for example, interaction confidence or Gene Ontology (Harris et al., 2004) terms annotated to the interacting proteins. In addition to the tools provided directly by databases, additional tools for graph-based interaction network analysis are provided by commercial and academic organizations, for example, Cytoscape (Shannon et al., 2003). Figure 1(a-e) shows the display of lsm7 (yeast) interactions in some visualization systems. Default settings have been used as much as possible.
Figure 1 (a) Visualization of lsm7 (yeast) interactions in DIP. (b) Visualization of lsm7 (yeast) interactions in Osprey. (c) Visualization of lsm7 (yeast) interactions in IntAct. (d) Visualization of lsm7 (yeast) interactions in MINT. (e) Visualization of lsm7 (yeast) interactions in BIND
Figure 1 (continued)
Figure 1 (continued)
The basic knowledge about protein interactions is currently increasing at a rapid pace, and a broad array of databases provides large data collections and powerful analysis tools, but the representation of protein interaction facts in near-text topic quality, taking into account interaction details such as protein state and dissociation constants, remains a challenge to the scientific community, both in terms of available tools and available data. Unlike other domains of molecular biology, in particular, DNA and protein sequences as well as macromolecular structures, protein interactions share the problem of data fragmentation in proteomics. Systematic deposition of data in public databases, and exchange of such data between databases, is only emerging. However, the standardization of interaction data through the PSI-MI standard, an increasing collaboration between protein interaction databases, and publicly available, open source analysis tools pave the way to public, user-friendly, and well-accessible protein interaction data resources, reflecting the huge biological significance of protein interactions.