1. Introduction
There is considerable heterogeneity in terms of both the numbers and types of cells present in different organs or tissues, reflecting their different biological functions. As an example, the cellular composition of the kidney comprises many diverse cell types including proximal tubule epithelial cells, distal tubule epithelial cells, collecting duct epithelial cells, endothelial cells, podocytes, mesangial cells, fibroblasts, and muscle cells. The proteome of any tissue, therefore, is actually a composite, reflecting the proteome of all its constituent cell types. The cellular makeup of tissues or organs can vary, however, not only during pathological states but also as a result of normal physiological changes. A prime example of this is the profound cellular changes that occur in the ovary and endometrium during the menstrual cycle. With diseases such as cancer, much of the normal tissue architecture may be lost with tumor areas consisting largely of malignant cells, but containing increased numbers of endothelial cells and infiltrating lymphocytes as part of the pathology. Additionally, within such a tumor and the surrounding tissue, cells that are at different stages of genetic evolution may be present, representing the transition from normal to frankly malignant and invasive pathology, and different microenvironments may be present, for example, normoxic and hypoxic areas.
Such tissue heterogeneity undoubtedly impacts on comparative studies, introducing more background “noise” to the analysis than would be present if the diseased cells alone were compared with their normal counterpart. The ability to isolate specific cell types or areas for experimental analysis would overcome this and generate more easily interpretable and readily applicable data. Conversely, however, while analysis of whole-tissue lysates may be more difficult to interpret, it does involve minimal manipulation of the in vivo state and the interplay between different elements of tissue, such as stromal-epithelial and endothelial-epithelial interactions, are undoubtedly important. The relative merits of either the whole-tissue approach or the selection of specific cells should be taken into account and the most appropriate method determined by the specific questions being addressed.
In terms of enriching for defined cell populations, various manual microdis-section, immunoisolation, or primary cell culture generation strategies have been explored. However, all these have inherent problems ranging from the laborious nature of the work to the possibility of introducing in vitro artifacts. One recent area of technological development that has facilitated the isolation of specific cell types or cellular areas without enzymatic digestion or cell culture is that of laser-based microdissection systems. Used extensively in experiments involving subsequent analysis of DNA or RNA from the isolated cells, the numbers of reports of successful use in proteomics-based studies are far fewer but nevertheless show the potential of the approach. The principles underlying the various laser-based microdissection instruments are outlined below, together with a consideration of the technical issues, and their use in various proteomics-based research illustrated.
2. Principles of laser-based microdissection systems
There are three main commercially available systems in current use. The first developed was the PixCell laser capture microdissection (LCM) system, which was developed by scientists at the National Institutes of Health in the United States (Emmert-Buck et al., 1996) and is marketed by the bioengineering company Arcturus. The system is based on a high-precision inverted microscope with a low energy, near-infrared laser mounted above it, a manipulator arm, and a computerized laser control and image capture platform. Using normal cleaned glass slides, the tissue section to be dissected is fixed and stained using appropriate protocols and the slide is placed on the microscope platform. A 6-mm-diameter Perspex cap, whose lower surface is covered with a transparent thermolabile ethylene vinyl acetate film, is then lowered into place onto the section with the film in direct contact with the tissue. By moving the stage so that the area of interest is in the field of view and by focusing the laser beam of adjustable diameter 7.5 to 30 |m through the cap onto an area containing cells of interest, the laser is then fired and the heat generated causes localized melting of the film and fusion with the selected cells directly below. The combination of relatively low energy and short-duration pulses (up to 5 ms) minimizes the heat damage to the tissue. Using the micromanipulator arm, the cap is then subsequently lifted from the section, complete with the fused dissected cells (Figure 1). Captured cells can then be solubilized using an appropriate extraction buffer either immediately or following further dissection and collection of cells. The most recently developed version of this system allows much of the process to be automated once appropriate areas of tissue are selected on the section image.
The PALM MicroBeam system couples “laser microbeam microdissection” (LMM) to ablate areas of tissue surrounding the cells of interest with “laser pressure catapulting” (LPC) to subsequently retrieve the selected area of cells (Schutze and Lahr, 1998). Again, the system is based around an inverted microscope to visualize the tissue section with the microdissection, that is, the movement of the stage and laser firing being automated. The operator delineates essentially the area containing the cells of interest on a computer screen and the process of ablation of the surrounding area is then computer-controlled. This strategy is therefore noncontact and nonheating, with cells of interest being cut round using cold photolysis generated by an ultraviolet N2 laser microbeam rather than positively targeted. The minimum width of the laser track separating adjacent areas is quoted as <1 |m, thus allowing quite selective ablation and delineation of infiltrating cells within areas of interest. To support the section and to allow subsequent visual examination of the retrieved material, a polyethylene naphthalate membrane backing is often used between the section and the glass slide. After microdissection, the laser is used to catapult the dissected tissue, complete with support membrane, into a collecting tube containing appropriate extraction buffer, a process termed laser pressure catapulting.
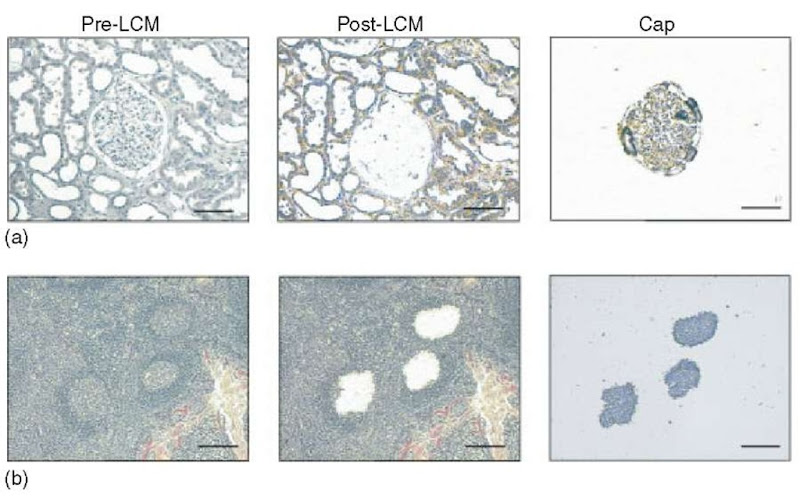
Figure 1 Examples of laser capture microdissection of (a) a renal glomerulus from kidney and (b) germinal centers from lymphoid tissue. Shown are the appearance of the tissue prior to and following capture and the captured cells present on the cap. The bar represents 100 |m
The most recently developed commercially available system is the Leica AS LMD. Similar in principle to the PALM system in that it employs a similar UV laser ablative microdissection strategy to remove surrounding tissue, it has a stated minimum diameter cut of <2.5 |m. However, the microscope is a conventional upright version and the slide is inverted such that the dissected material, once freed from surrounding tissue, is collected in a tube below the section, largely under the force of gravity (Kolble, 2000). Again the system is automated in that the operator delineates the dissection area, and in this case the stage remains still with the laser being moved and fired, a process controlled by the computer, to achieve the required ablation of surrounding areas.
3. Technical issues
The relative technical merits of the three systems with particular regard to resolution, ease of use and environment requirements, and sample visualization have been recently reviewed (Cornea and Mungenast, 2002). Particular issues pertaining to sample processing are briefly discussed below.
The predominant use of laser-based microdissection has been for the study of nucleic acids in selected cell populations, combined with a variety of downstream analytical techniques. The analysis of nucleic acids has the major advantage of requiring very little starting material due to the use of PCR amplification so that single-cell analysis is achievable. Additionally, paraffin-embedded, formalin-fixed material can often be used, allowing better visualization of the microdissection and providing access to a wealth of archival material. The uses of laser-based microdissection with nucleic acids have been reviewed and will not be covered further here (see Further reading).
As there is no amplification equivalent to PCR for proteins, proteomic analysis techniques that can be employed with microdissected material are dependent on a balance between their sensitivity and the time taken to accumulate sufficient material. For example, the dissection required to procure sufficient cells for large format 2D-PAGE requires tens of thousands of shots, which may represent several days of dissection, and there is a question, even with tissues that are relatively easy to dissect, as to whether, generally, only the most abundant protein species will therefore be examined. Those tissues requiring extensive microdissection will also inevitably result in a degree of compromise in terms of the purity achieved, and the degree of enrichment afforded in the protein profile relative to the starting material will also vary between tissue types (Craven et al., 2002).
Material fixed in cross-linking agents such as formalin is not suitable for proteomic investigation and hence frozen sections are used, although unfortunately of poorer morphology, particularly when viewed with inverted optics. Tissue sections need to be stained using protocols designed to allow good visualization but minimizing artifacts such as protein degradation and allowing subsequent analysis of extracted proteins. The initial “proof of principle” demonstration used a modified rapid hematoxylin and eosin staining method with proteinase inhibitors on frozen kidney and cervical tissue and showed that material collected by LCM could generate 2D-PAGE (see Article 22, Two-dimensional gel electrophoresis, Volume 5) protein profiles, with minimal effects of processing on either the profile or the subsequent mass spectrometric (MS; see Article 31, MS-based methods for identification of 2-DE-resolved proteins, Volume 5) sequencing results (Banks et al., 1999). Since this initial report, the examination of different tissue-processing regimens indicate that the type of fixative for the frozen sections is critical, with ethanol being the preferred option; various stains including hematoxylin and eosin, and methyl green and toluidine blue can be adapted and used successfully, as can the modified immunolabeling protocols using silver-enhanced gold labeling or fluorescent labeling (Craven et al., 2002; Mouledous et al., 2003a; Ahram et al., 2003). More recently, the process of “navigated LCM” has been used, which incorporates the use of fixed unstained sections for the microdissection, thus avoiding staining artifacts, but relies on images generated from adjacent stained coverslipped sections with consequent improved visualization of tissue architecture to delineate and guide the dissection process (Mouledous et al., 2003b).
4. Selected examples of applications in proteomics
The majority of published proteomics-based studies using laser-based microdissec-tion have utilized the PixCell LCM system unless indicated otherwise. The different studies are described below, subdivided on the basis of the subsequent proteomic techniques used to analyze the dissected material.
4.1. Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE)
One of the main separation techniques is 2D-PAGE. The use of 2D-PAGE,with protein lysates generated from the use of LCM provides a very encouraging endorsement of the approach, with the caveat that there are limitations with respect to sample and tissue type in terms of the degree of enrichment achieved (Craven et al., 2002). The initial demonstration of this combination of approaches showed the feasibility of using cells dissected from frozen tissue sections subjected to brief fixation and hematoxylin and eosin staining, with only modest effects of processing on the protein profile and compatibility with subsequent MS sequencing (see Article 31, MS-based methods for identification of 2-DE-resolved proteins, Volume 5) (Banks et al., 1999). As described previously, several other studies have also since explored other technical aspects of tissue processing. The main limiting factor with 2D-PAGE is the amount of material required, which may necessitate tens of thousands of laser shots and microdissection times of several hours or even days depending on the tissue type and still result in only a few hundred spots, that is, far less than normally visualized on large format gels. Inevitably, therefore, even with tissues that are relatively easy to dissect, only the most abundant protein species are likely to be examined.
In the first comparative analysis of microdissected normal versus malignant cells, analysis of lysates generated from approximately 50 000 cells from each matched sample from two patients with esophageal cancer allowed visualization of almost 700 proteins, with 98% similarity between normal and malignant protein profiles (Emmert-Buck et al., 2000). Ten proteins were tumor-specific with seven other proteins only found in normal epithelium, one of which was identified as annexin I. Using a combination of Western blotting of microdissected cell lysates and immunohistochemistry, an extension of this study confirmed complete loss or dramatic reduction of annexin I expression in both esophageal and prostate cancers, occurring even at the premalignant stage (Paweletz etal., 2000b). This study illustrates the use of a small number of gels requiring large amounts of microdissected protein as the initial step to determine differentially expressed proteins and then validating on a larger sample set using Western blotting with the necessity for much less material. A similar later study on prostate cancer found six tumor-specific changes in two cases examined, one of which was the tumor marker PSA, and which were only present in the microdissected epithelial compartments but not in stroma (Ornstein et al., 2000b). An extension of this study to an examination of 12 matched normal and high-grade tumor samples from 8 patients with cells obtained by LCM or manual microdissection and loading 100 000 to 140 000 cells per gel revealed 40 tumor-specific differences. However, disappointingly, only six of these were present in more than one case. Several of the proteins had been implicated before in tumor biology, for example, the 67-kDa laminin receptor and lactate dehydrogenase. (Ahram et al., 2002).
Similar success has also been seen with other cancers such as breast, ovarian, and pancreatic cancers. Examination of six matched pairs of normal breast ductal cells and ductal carcinoma in situ at either the whole tissue level or following LCM with up to 100 000 cells per gel identified 57 proteins to be differentially expressed and which met various criteria including being consistent between samples (Wulfkuhle et al., 2002). Only four proteins were identified by both whole tissue – and LCM-based approaches. Using a similar strategy, the 52-kDa FK506 binding protein, RhoGDI, and glyoxalase I were demonstrated to be uniquely overexpressed in invasive ovarian cancer compared with the low malignant potential ovarian tumors (Brown Jones et al., 2002), with downstream validation by a combination of Western blotting and reverse-phase protein arrays. In pancreatic cancer, microdissection and the subsequent 2D-PAGE analysis revealed about 800 spots per gel, with nine spots consistently differentially expressed including S100A6, trypsin, and annexin III (Shekouh et al., 2003).
The possibility of using fluorescence 2D-difference gel electrophoresis has been explored, which avoids the difficulties of gel-gel comparisons and allows more ready identification of spots from parallel gels for sequencing. Using limiting labeling, in a comparison of 250 000 cells from a matched tumour-normal esophageal sample, 1000 to 1100 protein spots were visualized (Zhou et al., 2002), with 58 proteins upregulated by > 3-fold in tumors and 107 proteins downregulated by > 3-fold. Using a matched SYPRO Ruby stained gel (see Article 27, Detecting protein posttranslational modifications using small molecule probes and multiwavelength imaging devices, Volume 5), annexin I was identified again as being downregulated and the tumor rejection antigen gp96 was upregulated, both of which were confirmed by Western blotting. Using a similar approach but with saturation labeling, normal murine intestinal epithelium and adenoma tissues from Min mice were compared (Kondo et al., 2003). Using only 2.7 to 6.6 |g of protein, which represented a 1-mm2 area of tissue dissected using the Leica LMD system from a 10-|m-thick frozen section with a collection time of 1 min, a 24-cm 2D-PAGE profile with 1500 spots was generated, 37 of which differed. Using a murine cell line run in parallel to generate sufficient material for MS analysis and subsequent Western blotting for validation, a number of proteins were identified including prohibitin, which is a putative tumor suppressor, HSP84, and a number of 14-3-3 zeta forms.
4.2. Western blotting
Western blotting of microdissected material provides not only information about the cellular localization of a protein but also confirmation of its molecular size, and, depending on antigen and antibody, requires relatively small amounts of material. Examples include the detection of actin, major vault protein, GAPDH, aldolase isoforms, pyruvate kinase, RhoGDI, annexin I, and gp96, with numbers of microdissected cells varying from approximately 250 to 40 000 (Craven and Banks, 2002; Unwin et al., 2003; Zhou et al., 2002; Paweletz et al., 2000b; Brown Jones et al., 2002; Wulfkuhle et al., 2002). Illustrating the potential of the approach not only in validating identified potential new markers but also in addressing questions regarding known proteins, microdissected material from benign and malignant prostatic epithelium was used to demonstrate that the vast majority of intracellular PSA exists in the free unbound form, but has the capability of binding to ai-antichymotrypsin form once outside the cell (Ornstein et al., 2000a).
4.3. Immunoassay
With the increasingly sensitive immunoassays employing chemiluminescent detection, laser-based microdissection represents a source of material for assay. Immunoassay of the amounts of the tumor marker prostate-specific antigen (PSA) in normal and malignant prostate epithelium using only several tens to hundreds of microdissected cells acquired in <15min, showed that malignant cells were found to contain up to sevenfold more PSA than normal epithelial cells, although there was overlap between the sets (Simone etal., 2000). Calculation of the actual number of PSA molecules per cell resulted in estimates of average values ranging from 10 000 to 1 000 000. More recently, hepatitis C virus core protein has been quantified by immunoassay in lysates prepared from microdissected liver cells prepared using the Leica LMD system and found to vary from 7 to 56pg/cell in infected samples, being more sensitive than immunohistochemistry (Sansonno et al., 2004).
4.4. Multiplex protein and antibody microarrays
The development of protein microarrays, Arraying proteins and antibodies, Volume 5, and Article 33, Basic techniques for the use of reverse phase protein microarrays for signal pathway profiling, Volume 5) enabling the simultaneous high-throughput profiling of many proteins in large numbers of samples, with only relatively small amounts of sample being required, is an area of considerable activity. Various formats exist including ones essentially analogous to cDNA arrays but with either antibodies or ligands arrayed onto which labeled tissue lysates can be applied. Alternatively, tissue lysates are arrayed, which are then probed with antibodies. It is increasingly evident that microdissected cell lysates are suitable for such applications.
Using a reverse-phase array approach, solubilized microdissected prostate tissue has been spotted onto nitrocellulose slides to generate microarrays containing up to 1000 tissue lysate spots/slide. Typically, these were produced using lysates resulting from only 500 to 3000 laser shots (equivalent to 2500-15000 cells). By using lysates from prostate cells covering the transition from normal epithelium through prostate intraepithelial neoplasia (PIN) to invasive cancer and by probing with a range of antibodies, changes during tumor progression were shown. These included decreased levels of cleaved poly(ADP-ribose) polymerase (PARP) and cleaved caspase-7, increased phosphorylation of Akt, but decreased phosphorylation of Erk and were interpreted as representing activation of prosurvival events (Paweletz et al., 2001). Results using antibodies against additional phosphorylated cell signaling molecules provided further evidence of activation of the Akt pathway (Grubb et al., 2003). A similarly designed study examining the activation status of several key signaling molecules involved in cell survival and proliferation in microdissected ovarian cancer also found changes in many of the variables examined but with generally no type or stage-specific patterns (Wulfkuhle et al., 2003). This may support the need to individualize patient samples in terms of the biological changes and the structuring of therapeutic strategies accordingly.
Antibody arrays have also been used to analyze microdissected samples. Using biotinylated extracts from 2500 to 3500 cells (approximately 0.5 | g) of both stromal and epithelial elements of squamous cell carcinoma of the oral cavity, together with an array containing 368 different antibodies against a variety of proteins, multiple protein differences were found with 11 proteins consistently changing in either relative levels or state of phosphorylation. Several of these were proteins involved in signal transduction pathways, with many exhibiting selective changes in either the epithelial cells or the stroma (Knezevic et al., 2001). This illustrates the importance of stromal-epithelial cell interactions in cancer.
4.5. Mass spectrometry
Although more conventionally used for protein identification on the basis of peptide mass fingerprints,matrix-assisted laser desorption ionization mass spectrometers equipped with time-of-flight detectors (MALDI-TOF; see Article 7, Time-of-flight mass spectrometry, Volume 5) are increasingly being used to generate profiles of intact proteins or fragments in complex biological samples in the context of disease-specific patterns, requiring only small amounts of protein. Using MALDI-TOF, direct analysis of 1250 cells from normal stroma, normal epithelium, carcinoma in situ, and invasive carcinoma prepared from a mastectomy sample and following addition of sinapinic acid to the LCM film immobilized on a standard stainless steel MALDI target generated distinct reproducible spectra containing 20-50 peaks from four cell types (Palmer-Toy et al., 2000). A subsequent investigation examined a range of factors and their effects on the quality of the profile generated, including section staining, ethanol treatment, and matrix application and showed that under optimal conditions, good quality profiles of at least the more abundant peaks could be generated from as few as 10 cells and again demonstrated that this approach could define distinct differences in profiles between normal and invasive breast epithelium (Xu et al., 2002).
Several studies have used the SELDI (surface-enhanced laser desorption and ionization) system developed by Ciphergen Biosystems (Fremont, CA, USA) to profile laser microdissected material. Based around a MALDI-TOF MS, proteins from complex mixtures are immobilized onto ProteinChip arrays containing 8-24 sample spots, by selective capture using the conventional chro-matographic chemistries of the chip surfaces. The first study to use SELDI-MS (surface-enhanced laser desorption and ionization-mass spectrometry) to analyze microdissected material was part of a larger study examining the specific forms of several prostate cancer markers in tissue samples and biological fluids. Using only 2000-3000 cells, several proteins were found to be differentially expressed between normal and malignant samples with a peak detected only in the prostate carcinoma samples being found to have the same molecular weight as purified prostate-specific membrane antigen (PSMA) (Wright et al., 1999). Several studies have now shown that microdissected material can be examined in this way to discriminate between normal and diseased cells in a variety of cell types and diseases, although mainly cancers (Paweletz etal., 2000a; von Eggeling etal., 2000; Batorfi etal., 2003; Zheng et al., 2003; Melle et al., 2003). The numbers of peaks generated in a sample vary with ProteinChip type, extraction buffer, and tissue and although reproducible protein “fingerprints” can be generated using as few as 25-100 cells, far more proteins can be seen with increasing loads and in most cases between 500 and 10 000 cells have been used, the amount of protein present in these dissections being very dependent on factors such as cell type and section thickness however. Although in a number of cases, particular discriminatory peaks have been found, the challenge now lies in being able to identify the particular proteins or protein fragments that will provide further information. One approach that was used successfully to identify a peak of approximately 36 kDa as annexin V, which discriminated between normal pharyngeal epithelium and head and neck squamous cell carcinoma was that of subsequent 2D-PAGE and MS sequencing (Melle et al., 2003). This was possible due to the size of this particular peak, making it amenable to such separation. However, for smaller proteins or fragments, this is still a challenge.
5. Summary
The number of studies using laser-based microdissection methods as a prelude to subsequent proteomic analyses is still small, but sufficient to illustrate the promise of such technology in overcoming the problems of tissue heterogeneity. The use of highly sensitive downstream analysis techniques that require relatively small amounts of sample, such as protein and antibody microarrays and MS profiling are likely to be the main areas in which such microdissection techniques will contribute more extensively in the future. The combination with multidimensional liquid chromatography tandem MS (see Article 13, Multidimensional liquid chromatography tandem mass spectrometry for biological discovery, Volume 5) is also a possibility, although not yet explored. The analysis of different cell populations in abnormal tissues will clarify the molecular basis of changes occurring in disease and during progression, and in defining the important role of interactions between the different cellular compartments. Ultimately, in addition to increasing our understanding of the underlying disease pathogenesis, this has the potential to contribute in various areas of clinical utility ranging from marker discovery for diagnosis and prognosis, to identification of specific disease-related alterations in individuals and appropriate tailoring of targeted therapy.