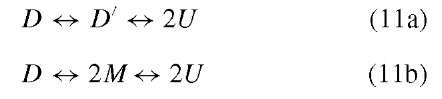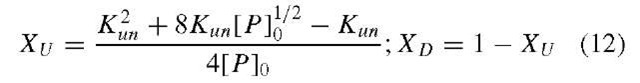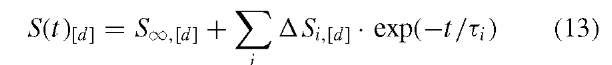A. General Experimental Strategies
As discussed above, experimental studies of protein folding reactions fall into the category of either equilibrium or kinetics studies, with the former yielding thermodynamic information about the energy differences between the native and denatured structural states and the latter studies providing information about the folding pathway and the height of energy barriers between important species on this pathway. In general, to perform either an equilibrium (thermodynamics) or time-dependent (kinetics) study, one must be able to experimentally monitor a signal that tracks the population of the structural states of the protein.
There are a number of ways this can be done. The most convenient experimental methods involve solution-phase spectroscopic measurements; among these methods are absorption spectroscopy, fluorescence, circular dichroism, and nuclear magnetic resonance. Other methods include differential scanning calorimetry, light scattering, elec-trophoresis, and chromatography. This section gives a brief description of the advantages and disadvantages of some of the above methods. These methods are not equally applicable to equilibrium and time-dependent studies of protein unfolding, as some methods have a rapid response and some have a slow response. Methods also differ in their intrinsic sensitivity, which is related to the concentration of protein necessary to perform the measurement, their ease and economy of use, and whether they provide auxiliary information about the structure of the protein in its native and denatured states. What is meant by the last statement is that some of the spectroscopic signals can provide information about the secondary or tertiary structure of the protein species. For most types of spectroscopy, the signal arises from particular amino acid residues (e.g., aromatic side chains or peptide bond), thus differences in the signals for the conformational states can be related to differences in the local environment of these amino acid residues (e.g., tryptophan residue 140 in staphylococcal nuclease; see Fig. 1). If there are only a very few of such signal origination sites, then site-specific information can be obtained. If there are many probe sites and they are distributed throughout the protein’s structure, then the method yields global information (e.g., signal from the amide linkage in the peptide backbone; see Fig. 1). It goes without saying that the protein sample to be studied must be well defined with regard to purity, and solution conditions must be selected and controlled to be relevant to other functional studies and studies with other proteins. Neutral pH, 20°C, and an ionic strength of 0.1 to 0.2 are the most commonly employed solution conditions.
A key to most of these methods and their use in protein unfolding studies is that the signal is a mole-fraction weighted average of the signals of each protein species. That is, for the simplest case of a thermodynamics study of the transition between a native, N, and unfolded, U, state of a protein, the observed signal, S, can be expressed as:
where Xi is the mole fraction of each species i and Si is the intrinsic signal of species i. This relationship applies to most solution optical spectroscopic methods. Clearly, for a particular spectroscopic signal to be useful for tracking a N ^ U transition, the signal of the N and U states must be sufficiently different. The native (XN ) and unfolded (XU ) mole fractions are directly related to the equilibrium constant in Eq. (2), as:
The transition from the native state to the unfolded state, or vice versa, can be induced in several ways, essentially by varying the solution conditions in a way that changes the equilibrium between the native and unfolded state. The transition may be induced by varying temperature, adding chemical (chaotropic agent) denaturant, adding acid or base, or increasing pressure. In the case of multimeric proteins, subunit dissociation, which may be accompanied by denaturation of the subunits, can be induced by dilution of the protein. Before discussing the various spectroscopic methods, some thermodynamic relationships are presented for describing the transitions induced in the above ways.
B. Basic Thermodynamic Relationships
Table I gives some widely accepted relationships for describing the variation of AG°un for a two-state N ^ U transition with temperature, chemical denaturant, pH, or pressure as the perturbations. One of the equations in Table I, when combined with those above and Eqs. (13), can be used to describe data as a function of the denaturing condition. The thermodynamic parameters related to the relationships in Table I are briefly described below.
1. Thermal unfolding: A Hn and A SUn are the enthalpy and entropy changes for a two-state unfolding reaction. Both AHuon and ASuon may be temperature dependent, when the heat capacity change, A Cp, has a nonzero value. In this case, Eq. (7b) in Table I (the Gibbs-Helmholtz equation) should be used, where the AHoun and AS°o un are values at some defined reference temperature, To (e.g., 0° or 20°C).6′ 7 The heat capacity change for unfolding of proteins is typically found to be positive and to be related to the increase in solvent exposure of apolar side chains upon unfolding. That is, a positive A Cp is a result of the hydrophobic effect. A consequence is that the AG°un(T ) for unfolding ofa protein will havea parabolic dependence on temperature and will show both high-temperature and low-temperature induced unfolding.8
2. Denaturant-induced unfolding: The empirical relationship in Table I for chemical denaturation includes
AGo,un, the free energy change for unfolding in the absence of denaturant, and m, the denaturant susceptibility parameter (= SAGun/S[d]), where [d] is the molar concentration of added chemical denaturant.9’10 Through an empirical relationship, the given equation appears to adequately describe the pattern for denaturant-induced unfolding of a number of proteins. The AG°oun value is a direct measure of the stability of a protein at the ambient solvent conditions, which can be moderate temperature and pH (e.g., 20oC and pH 7). The m value also provides structural insights, as m values have been suggested to correlate with the change in solvent accessible apolar surface area upon unfolding of a protein.11 For example, a relatively large m value (i.e., a high susceptibility of the unfolding reaction to denaturant concentration) indicates that there is a large change in the exposure of apolar side chains on unfolding, which might be the case for a protein that has an extensive core of apolar side chains that are exposed upon denaturation.
3. Acid-induced unfolding: The relationship for acid-induced unfolding assumes that there are n equivalent acid dissociating groups on a protein that all have the same pKa,U in the unfolded state and that they are all perturbed to have a pKa,N in the N state. If the pKa,N is more than 2 pH units lower than pKay, then the equation simplifies with the denominator of the right term going to unity. The simplest relationship for acid-induced unfolding includes AG°oun, the free energy of unfolding at neutral pH; n, the number of perturbed acid dissociating residues; and their pKa U in the unfolded state. Presumably, n should be an integer and pKa^ U should be approximately equal to the values for such amino acids as glutamate, aspartate (e.g., pKa’ U should be about 4 to 4.3) or histidine (e.g., pKa’U should be around 6.5).
4. Pressure-induced unfolding: In the relationship for pressure, P, induced unfolding of proteins, AG°ouun is again the value of the free energy change at 1 atmosphere pressure and A Vun = VU — VN is the difference in volume of theunfolded and native states. Pressure-induced unfolding studies require a specialized high pressure cell.12 • 13
TABLE I Relationships Describing Two-State Transitions in Proteins
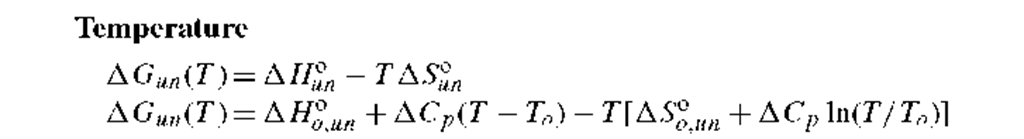 |
(7a) (7b) |
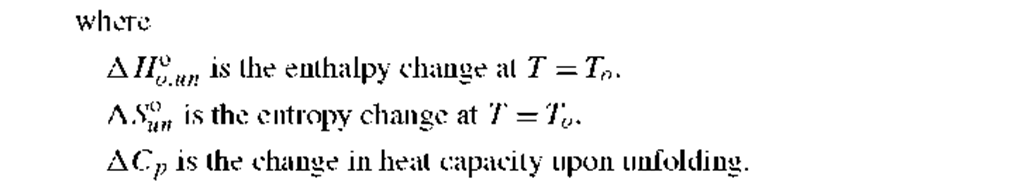 |
|
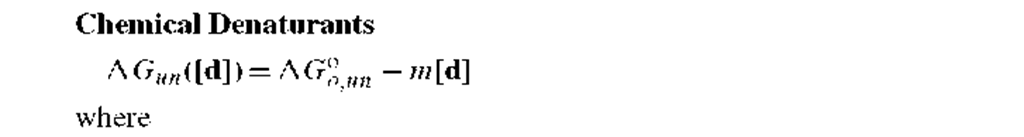 |
(linear extrapolation model) (8) |
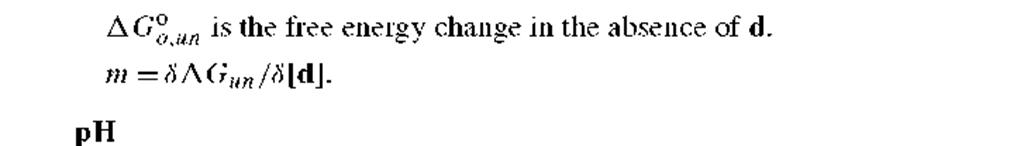 |
|
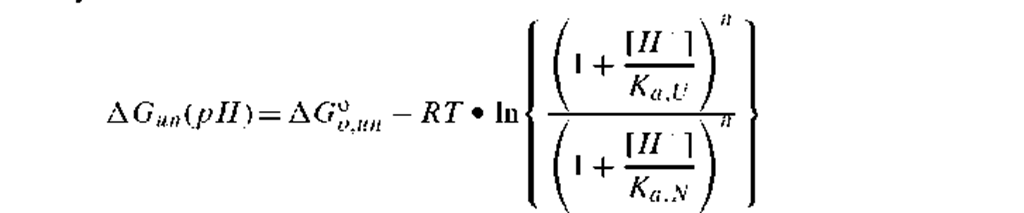 |
(9) |
 |
|
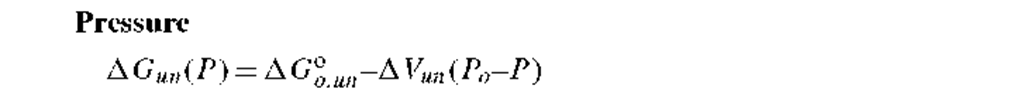 |
(10) |
 |
For a two-state transition, .![]() for the unfolding of a native, N, to an unfolded, U, state of a protein) the mole fractions of the N and U states are given as
for the unfolding of a native, N, to an unfolded, U, state of a protein) the mole fractions of the N and U states are given as![]() when
when![]() and the function for
and the function for![]() is taken from above the average fluorescence signal,
is taken from above the average fluorescence signal,![]() where x is a generalized perturbant.
where x is a generalized perturbant.
5. Dissociation/unfoldingofoligomericproteins: Olig-omeric proteins are interesting as models for understanding intermolecular protein-protein interactions. A general question for oligomeric proteins, including the simplest dimeric (D) proteins, is whether the protein unfolds in a two-state manner, D 2U, or whether there is an intermediate state, which might be either an altered dimeric state, D’, or a folded (or partially folded) monomer species, M. Models for these two situations are as follows:
For a D ^ 2U model, the relationships between the observed spectroscopic signal, Sexp; the mole fraction of dimer, XD , and unfolded monomer, XU ; and the unfolding equilibrium constant (Kun = [U]2 /[D]) will be given by Eq. (5) and
where [P]0 is the total protein concentration (expressed as monomeric form), where Si is the relative signal of species i and where Kun will depend on the perturbant as given by one of the above equations. That is, the transition should depend on the total subunit concentration, [P]0, and on any other perturbation axis.
C. Experimental Signals
1. Absorbance Spectroscopy
Absorbance spectroscopy (difference spectroscopy) monitors conformational transitions in macromolecules by measuring absorbance changes, usually in the aromatic region of the ultraviolet (UV) spectrum. The amino acids tryptophan and tyrosine are the most important chromophores in the UV region for proteins. As mentioned earlier, tryptophan residues are often engineered into proteins as reporters of local and/or global environment.
The indole ring of tryptophan and the phenol ring of tyrosine show sensitivity of their absorbance spectrum to solvent polarity. There is a blue shift in the absorbance of indole and phenol upon increasing solvent polarity. As a result, there will often be a blue shift in the absorbance of tryptophan (typically monitored as a decrease in absorbance in the 291- to 294-nm region of the spectrum) or tyrosine (at 285 to 288 nm) upon unfolding of a protein and a consequent increase in the exposure of these aromatic side chains to water.14 Tryptophan’s absorbance is also sensitive to the local electrostatic field; changes in indole-charge interactions can cause either red or blue shifts upon protein unfolding.15
Table II gives the typical concentration range used for unfolding studies with proteins using this and other methods. The sensitivity of difference absorbance measurements will depend on the molar extinction coefficient of the chromophore and their number, but a concentration range of 0.01 to 0.1 mM protein is usually needed for reasonable signal to noise with a 1-cm pathlength cell. Thermal scans, to induce the unfolding transition, are easy to perform with accessories available for most absorbance spectrophotometers. Chemical denaturant- or pH-induced transitions can be less convenient (unless one has automated titration equipment), since a series of solutions with equal protein concentration and varying denaturant must be prepared. With any of these perturbing conditions, it is important to realize that the variation in the conditions itself (i.e., varying temperature, pH, chemical composition) can lead to a "baseline" change in the absorbance signal from the native and unfolded species.16 So long as these baseline trends are linear and not as large as the ab-sorbance change associated with the conformational transition, the baseline trends can be corrected for in the data analysis.
The advantages of absorbance measurements are the ready availability, ease of use, and low cost of the instrumentation. The biggest disadvantage is that it is less sensitive than some other methods.
2. Circular Dichroism
Circular dichroism (CD) is a very commonly used method for studying protein conformational changes. The far UV spectral region (180 to 250 nm) is dominated by ab-sorbance by peptide bonds, and there are signature spectra for a-helix and other types of secondary structure in a protein. Additionally, the aromatic CD spectral region of 250 to 300 nm senses the chirality around the aromatic amino acid side chains and there is usually a structured aromatic CD spectrum for the native state of a protein.14, 17, 18
The effective sensitivity of CD is comparable to or slightly better than that of difference UV absorbance spectroscopy. CD instruments can be purchased with thermoelectric cell holders for thermal scans and with automated titrator syringe pumps for chemical denaturant titrations. Since the far-UV spectral regions is important in protein unfolding studies, it is necessary to work with salts and buffers that have minimal absorbance in this region. When performing CD measurements, it is necessary to pay attention to the buffer and salts and other solution components (e.g., chemical denaturants) being used, particularly if one wishes to make measurements below 200 nm, as various buffers, salts, and denaturants can absorb a significant amount of light in the far-UV. Schmid14 has provided a number of practical tips regarding the application of CD for studies with proteins. There is less interference by buffer, salts, etc. in the aromatic UV spectral region. Whereas the aromatic CD signals can sense the loss of tertiary structure in a protein as it denatures, the CD signals in this region are much smaller than those in the far-UV CD region, giving a lower signal-to-noise ratio. Baseline slopes, as one varies temperature or chemical denaturant, also must be considered in CD measurements in both the far-UV and aromatic spectral region; however, the baselines trends are usually not large.
TABLE II Solution Methods for Monitoring the Progress of Protein Unfolding Transitions
|
Method |
Conc. Range (m Mf |
Structure Sensed |
Kinetic Applications |
|
|
Absorbance |
0.01-1 |
TS/AT |
Local |
*** |
|
Circular dichroism |
0.01-0.1 |
TS/AT |
Secondary |
*** |
|
Fluorescence |
0.0001-0.01 |
TS/AT |
Local/tertiary |
*** |
|
FTIR |
0.5-2 |
TS |
Secondary |
* |
|
Light scattering |
0.1-1 |
No |
Size and shape |
* |
|
NMR |
1 -10 |
No |
Local/tertiary |
* |
|
DSC |
0.02-0.2 |
TS |
Tertiary |
e |
|
Activity/binding |
_ft |
P |
Tertiary |
* |
|
Chemical reactivity |
variable |
P |
Local/tertiary |
* |
|
Chromatography |
c |
No |
Size and shape |
— |
|
Electrophoresis |
c |
Gradients |
Size, shape, charge |
— |
|
Potentiometry |
0.1-1 |
No |
Local |
— |
a Concentration ranges are for typical experiments with a 20-kDa protein.
b The concentration range will depend on the method being used to measure enzymatic activity or ligand binding.
c The concentration of protein varies during the course of the experiment as the sample flows through the column, gel, or capillary. Initial concentrations are usually in the range of 1 mg/mL.
d "TS" refers to the ability to perform thermal scans to unfold a protein; "AT" refers to the ability to perform automated titrations of a protein sample with chemical denaturant, acid, or base while the sample is loaded in the instrument. The label "P" indicates that an automated thermal scan or titration may be possible for certain applications, though this is not commonly done. The "Structure Sensed" column lists the features of the protein structure (e.g., secondary and tertiary structure, local interactions, etc.) that are sensed by the method. Some of these entries are judgment calls. The "Kinetic Applications" column indicates the amenability of the method to protein folding/unfolding kinetics experiments. A label "***" indicates that transient mixing or other means are available for the rapid initiation of the reaction. A label "*" indicates that the method is amenable to study relatively slow reactions (i.e., by a hand-mixing experiment).
e Through variation of thermal scan rate or a frequency domain application of DSC, it is possible to obtain kinetics information.
A difference between far-UV CD and other optical methods is that CD signals observe changes throughout the structure of the protein (i.e., its secondary structure) and the magnitude and direction of the signal changes can be more directly related to changes in structure (e.g., a loss of ellipticity at 222 nm can be related to a loss of a-helix).
3. Fluorescence
Fluorescence is the most sensitive of the commonly used optical methods for studying protein unfolding transitions.14′ 19—21 The absolute sensitivity depends on a number of factors (e.g., lamp or laser intensity, cell pathlength, chromophore extinction coefficient, and quantum yield), of course, but commercial fluorometers can usually detect signals down to the 10-nM range. Either intrinsic or extrinsic fluorophores can be used. The most commonly used intrinsic fluorophores are the tryp-tophan and tyrosine residues, with the former being the most important due to its larger molar extinction coefficient and a redder absorbance and emission. The fluorescence of tryptophan residues is very dependent on the local microenvironment of its indole side chain, making tryptophan fluorescence responsive to the structure of a protein. This spectral responsiveness is in terms of its emission maximum and its quantum yield. For example, the emission maximum of tryptophan almost always shifts to longer wavelengths (red shifts) upon unfolding a protein and increasing the solvent exposure of this amino acid side chain. There is a large literature about the fluorescence of tryptophan residues in proteins and its use to study changes in the structure of proteins.19
A variety of extrinsic fluorophores can be attached to proteins to serve as fluorescence probes. These can be selected to maximize sensitivity and to avoid contamination (i.e., by moving to longer absorption and emission wavelengths) from other absorbing components.22 With both intrinsic and extrinsic fluorescence probes, the method focuses only on these probes sites, which might be as few as a single site on a protein.
Like the signals from absorption spectroscopy and CD, fluorescence intensity signals (either at a single wavelength or integrated over the emission envelope) follows Eq. (5) and can be used to extract thermodynamic information. However, there are other easily measured fluorescence signals (emission maximum and anisotropy) that do not follow the mole fraction averaging of Eq. (5).19 The apparent emission maximum of a protein will be dominated by the structural state, native or unfolded, which has the higher quantum yield. Consequently, the apparent emission maximum will frequently not give a true reflection of the population of native and unfolded states, thus limiting the value of this type of fluorescence measurement for use in recovering thermodynamic parameters. (Rather than use the apparent emission maximum, it is better to perform curve fitting with composite spectra of the native and unfolded states.)
Fluorescence anisotropy values for the fluorescence of a fluorophore on a protein will depend on the fluorophore’s rotational freedom and fluorescence lifetime. Because the motional freedom of intrinsic or extrinsic fluorophores will usually increase when a protein unfolds, a change in a protein’s fluorescence anisotropy is expected upon unfolding. However, to properly use anisotropy to analyze the thermodynamics (or kinetics) of an unfolding transition, Eq. (1) should be replaced with one that includes the fluorescence quantum yield of the protein’s structural states (see Reference 19).
As with the above-listed optical methods, fluorescence instruments are designed to allow automated thermal scans and/or titrations. The baseline problem can be more significant with fluorescence than the other methods and should not be ignored. In particular, it is well known that the fluorescence intensity of fluorophores will decrease with increasing temperature, regardless of whether there is a conformational transition. While baseline trends may not be linear over extensive ranges of the perturbing variable (e.g., temperature or chemical denaturant), it is usually adequate to assume linear slopes over a limited range of the variable.
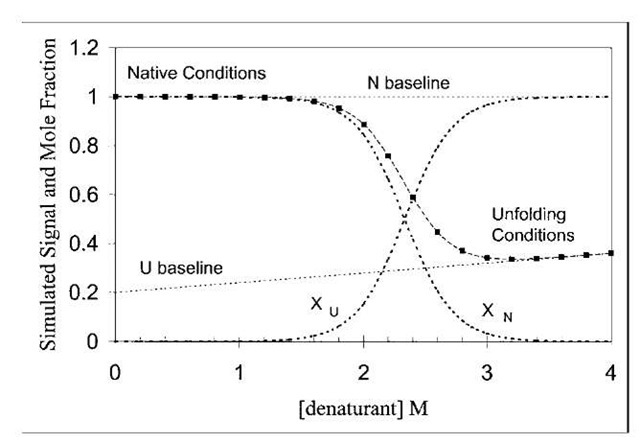
The advantages of fluorescence for studying protein unfolding reactions are the wide concentration range that can be measured and the responsiveness of the signal to the microenvironment of the fluorophore. Additionally, fluorescence signals of the native and unfolded state can provide a modicum of structural information about these states (at least with respect to the microenvironment of the fluorophores). Figure 3 shows simulated data for the denaturant-induced unfolding of a protein, as would be monitored by fluorescence intensity measurements.
4. Differential Scanning Calorimetry
Another frequently used method is differential scanning calorimetry (DSC), which measures the variation in the specific heat of a protein containing solution as a protein is thermally unfolded.23—25 As opposed to the above optical techniques, where photons are being measured, calorimetry measures the transfer of heat associated with the thermally induced conformational transition. DSC and related types of calorimetry are intrinsically less sensitive than the optical methods. Nonetheless, advances in the technique have made it possible to perform DSC studies with samples as low as 0.1 mg/mL.
Temperature is scanned in DSC measurements, so it is the variable that causes the structural transition of a protein. DSC data are typically presented as thermograms that yield a heat-capacity maximum corresponding to the thermal transition temperature, TG , and an enthalpy change, A Hun, for the transition. The A Hun value can be determined either by integration of the thermogram or by curve fitting (i.e., fitting a van’t Hoff equation to the shape of the thermogram). Referring to these two A Hun estimates as the calorimetric and van’t Hoff values, the ratio of the calorimetric and van’t Hoff AHun values can be used to determine whether the transition is best described as a two-state process. That is, a ratio of 1.0 (indicating that two AHun estimates are essentially the same) means that the structural transition is two state.
FIGURE 3 Simulation of denaturant-induced unfolding of a protein in a two-state manner. A simulated fluorescence signal (■) is plotted vs. denaturant concentration for a protein, using Eq. (8). The simulated fluorescence signal decreases with addition of denaturant because the unfolded species has a smaller fluorescence signal (fluorescence i s quenched on unfolding). The pre- and post-transition baselines may slope, as shown. The fraction of unfolded species (XU ) increases from left to right as the fraction of native species (XN) decreases.
5. Nuclear Magnetic Resonance
Nuclear magnetic resonance (NMR) spectroscopy is a powerful method for studies with proteins, as there is such a large number of resolved signals (due to the individual nuclei, such as the 1H and 13C atoms in the backbone and/or side chains of the amino acids).26 • 27 This gives the potential to track conformational transitions by observing changes at a large number of individual sites on the protein. This is further made possible by the fact that the signals (peaks having various chemical shifts) are usually widely dispersed in the native state of a protein, as a consequence of the sensitivity of the resonance peak for individual nuclei to the local magnetic field, which in turn is related to the three-dimensional structure of the protein. Unfolded proteins, by comparison, usually have a much narrower range of resonance peaks for similar amino acid components.
Tracking any of the individual resonance signals, such as those assigned to histidine or tryptophan residues, as a function of denaturing condition (e.g., temperature, pH, or added chemical denaturant) provides a way to study the unfolding process, as the signal is transformed from that of the native state to that of the unfolded state. An important difference between NMR and the above optical methods is that NMR signals can be dynamically averaged signals or individual signals can appear for the native and unfolded states. The latter results if the rate of interconversion of the conformational states is relatively slow in comparison to the difference in resonance frequencies of the signals for the two states.
The use of NMR for proteins studies is usually limited to proteins having molecular weight of about 25 kDa or less. The method requires a relatively high concentration of protein, compared to other methods.
Besides the above application of NMR to track the population of native and unfolded states, NMR also can provide very high-quality information about the tertiary and secondary structure of proteins. In addition, pulsed isotope labeling experiments can provide information about the pathway for protein folding reactions and can provide estimates of the unfolding equilibrium constant at individual sites on the protein.26 27
6. Other Experimental Methods
Fourier transform infrared (FTIR) vibrational spec-troscopy senses the hydrogen bonding pattern of the peptide bonds of a protein and can detect unfolding transitions in terms of changes in the secondary structure patterns.28 As compared to CD, which also senses secondary structure, FTIR is relatively more responsive to j-sheet structures. A disadvantage of FTIR is that it requires a higher protein concentration and that it is more difficult to automate for titration experiments.
Light-scattering methods, such as small-angle X-ray scattering, or quasi-elastic light scattering, can provide information about the size of a protein, in terms of its radius of gyration. Unfolding or aggregation reactions are detected as increases in the hydrodynamic radius.29 These scattering methods are also relatively difficult to adapt to temperature or titration experiments.
The ability of a protein to bind a specific ligand or to have catalytic activity can be used to determine the population of native species. The possibilities are numerous, depending on the way that activity and binding are measured. These activity/binding assays should be easy to automate for a series of denaturant concentrations or pH values.
Size exclusion chromatography and gel or capillary electrophoresis are methods that separate protein molecules based on size (or size and charge).30-32 In these methods, the protein sample travels down the column, gel slab, or capillary and, for a pure protein, should exit as a single peak traveling past the detector. Denatured proteins should appear to have a larger hydrodynamic radius and should travel more slowly. If the kinetics of inter-conversion of the native and unfolded species is slower than the time needed to travel through the column (gel or capillary), then it is possible to detect individual peaks for the native and unfolded species. If the interconver-sion is rapid, a kinetically averaged peak position will be observed.
An example of a potentiometric measurement is one in which the pH (or number of protons bound versus those bound at some reference condition) is measured as a function of the denaturing condition. Such an approach would require a difference in the pKa of one or more amino acid side chains in the native and unfolded state. Usually, several such amino acid side chains are in a protein. However, the potentiometric approach requires technical skill, and it is difficult to use in combination with high concentrations of chemical denaturants or temperatures far from ambient.
D. Kinetics Experiments
Studying the kinetics of folding or unfolding a protein involves the rapid initiation of the reaction by either removal of the denaturing condition (to initiate folding) or addition of the denaturing condition (unfolding). The most common way in which this is done is by a stopped-flow mixing device, in which a solution of the protein in one solution condition (e.g., neutral buffer with no chemical denatu-rant) is rapidly mixed with a second solution (e.g., concentrated chemical denaturant or acid). Temperature or pressure can also be used as the perturbing condition. Upward temperature jumps can be initiated by high-powered laser pulses or electrical discharges in the solution. Downward pressure jumps can be initiated by releasing some type of valve after high pressure has been established. Other ways to rapidly initiate a protein folding or unfolding reaction include such things as laser flash-induced chemical reactions, which can dissociate heme-carbon monoxide bonds of heme proteins. The remaining discussion will emphasize stopped-flow mixing reactions, since these are the most widely available approaches.
For small, globular proteins, the kinetics of denaturant-induced or acid-induced unfolding reactions are often found to be described as a mono-exponential process, indicating that there is a single energy barrier between the native and unfolded species. For larger proteins, particularly those with multiple domain structures, one often finds the kinetics to be more complicated. In those cases, the folding or unfolding reaction may be described by more than one exponential decay term. This can be an indication of the existence of multiple, slowly intercon-verting unfolded states, the existence of multiple energy barriers along the folding/unfolding pathway, the existence of more than one pathway, or the existence of some off-path (or dead-end) species. The challenge in kinetics experiments is to first determine the minimum number of decay terms needed to describe the reaction and to then determine a reaction mechanism consistent with the kinetics data. Often, unique mechanisms cannot be determined and it is the art of the scientist to establish which mechanism is most reasonable for the system being studied. In many cases research will focus on determining the number of intermediates on the folding pathway and in trying to gain structural information about these intermediates.
Whether studying folding or unfolding, the reaction should be described with the following general empirical relationship:33 34
where S(t)[d] is a time-dependent change in some generalized signal at denaturing condition described by [d], Sx , [d], is the signal at equilibrium (infinite time after initiation of reaction) at the denaturing condition, and ASi , [d] is the signal amplitude associated with relaxation time constant, Ti . If there is only a single step in the process (i = 1), the reaction will be a mono-exponential. With more steps, the reaction becomes bi-, tri-, … exponential, and fitting the above equation to the data can be difficult. Even with the number of terms and the corresponding values of Ti determined, it is still a challenge to relate the ti and the associated amplitudes to a reaction mechanism. How this can be done is beyond the scope of this entry and we refer readers to References 35 and 36.
To experimentally apply the above equation, it is necessary to monitor some signal that responds to the structural state of the protein, just as is the case in equilibrium studies. For very rapid folding/unfolding reactions, the monitoring method must itself be able to respond more rapidly (with adequate signal to noise) than the chemical reaction. Fluorescence, absorbance, and circular dichroism are fairly rapidly responding optical methods and enjoy widespread use in combination with stopped-flow mixers for this purpose. As indicated in Table II, some of the other methods are not easily adapted for the rapid initiation and continuous monitoring of a reaction’s progress. Obviously, when a reaction occurs on the time scale of minutes to hours, then a hand-mixing experiment can suffice.