Transcription is a highly complex process because of its defined initiation and termination sites in the genome and the subsequent processing and regulation of its synthesis. The steady-state level of a protein in the cell is the balance of its rate of synthesis and degradation. The synthesis is determined primarily by the steady-state level of its mRNA. Thus, the rate of transcription often determines the level of its gene product in vivo.
As mentioned earlier, RNA synthesis is catalyzed by the RNA polymerase in all organisms. Prokaryotes express a single RNA polymerase used for synthesis of all RNAs, while eukaryotes encode multiple RNA polymerases with dedicated functions. RNA polymerase I (Pol I) in eukary-otic cells is responsible for synthesis of ribosomal RNA, which accounts for more than 70% of total RNA in the cell. Pol III catalyzes synthesis of small RNA molecules, including transfer RNAs which bring in appropriate amino acids to the ribosome for protein synthesis by using their "anti-codon" triplet bases. Pol II is responsible for synthesis of all other RNA, specifically mRNA.
RNA polymerases of all organisms are complex machines consisting of multiple subunits which alter conformation. A variety of structural analyses show the presence of a 2.5-nm-wide "channel" on the surface of all DNA polymerases which could be the path for DNA. The RNA polymerase holoenzyme binds to a promoter-specific recognition sequence upstream (5′ side of the transcribed strand) of the site of synthesis initiation. While the RNA polymerase is normally present as a closed complex with nonspecific DNA, in which DNA base pairs are not broken, a significant conformational change produces the open complex when RNA the enzyme binds the promoter, unwinds the DNA duplex, and is poised to initiate RNA synthesis.
As in the replication process, initiation is the first stage in transcription and denotes the formation of first phospho-diester bond. Unlike in the case of DNA synthesis, RNA chains are initiated de novo without the need of a primer. However, when a primer oligonucleotide is present, RNA polymerases can also extend the primer as dictated by the template strand. A purine nucleotide invariably starts the RNA chains in both prokaryotes and eukary-otes, and the overall rate of chain growth is about 40 nucleotides per second at 37°C in E. coli. This rate is much slower than that for DNA chain elongation (~800 base pairs per second at 37° for the E. coli genome).
RNA synthesis is not monotonic, and RNA polymerases can move backward like DNA polymerases do for their editing function in which an incorrectly inserted deoxynu-cleotide is removed by 3′ exonuclease activity. RNA poly-merases stall, back track, and then cleave off multiple newly inserted nucleotides at the 3′ terminus. Subsequently, polymerases move forward along the DNA template and resynthesize the cleaved region. Based on the segment of DNA covered by an RNA polymerase as analyzed by DNA footprinting, it has been proposed that the enzyme alternatively compresses and extends in its binding to the DNA template and acts like an inchworm in its transit.
RNA polymerases of both prokaryotes and eukaryotes function as complexes consisting of a number of subunits. The E. coli RNA polymerase enzyme with a total molecular mass of about 465 kD contains two a-subunits, one j-and one j’-subunit each, and a a-subunit which provides promoter specificity. During chain elongation, a ternary complex of macromolecules among DNA template, RNA polymerase, and nascent RNA is maintained in which most of the nascent RNA molecule is present in a single-stranded unpaired form. The stability of the complex is maintained by about nine base pairs between RNA and the transcribed (noncoding) DNA strand at the growing point.
While DNA replication warrants permanent unwinding of the parental duplex DNA, asymmetric copying of only one strand by RNA polymerase requires localized strand separation which is induced by the polymerase itself, resulting in a transcription bubble. During chain elongation, this bubble moves along the DNA duplex. Initiation of RNA synthesis is enhanced in an in vitro reaction with supercoiled duplex circular DNA template in which base pairs are destabilized due to torsional stress. Unwinding of the helix at the transcription site causes overwinding (positive supercoiling) of the template DNA ahead of the transcription bubble and underwinding (negative super-coiling) behind the bubble.
A. Recognition of Prokaryotic Promoters and Role of -Factors
In prokaryotic RNA polymerases, the a -factor is required for promoter recognition and binding. It is loosely bound to the core complex and released after the nascent RNA chain becomes 8-9 nucleotides long. The core polymerase with a-factor has a high affinity for nonspecific DNA. The a -factor alters the conformation of theholoenzyme so that its affinity for nonspecific DNA is reduced and the specific binding affinity for the promoter is significantly enhanced.
More than one type of a-factor is present in E. coli, and more such factors are present in other bacteria. These different factors may have specialized functions in altered growth conditions, cause a global change in transcrip-tional initiation dueto their recognition of distinct -35 and -10 sequence elements, and have a preference for different promoters.
RNA chain termination in bacteria occurs by two mechanisms, one with assistance of a protein factor rho (p) and the other without need of a protein. In both cases, termination occurs at a specific terminator sequence in the gene, at which the RNA polymerase stops adding nucleotides to the growing RNA chain, which is then released from the template. The terminator sequence often has a "hairpin" structure which results from intramolecular base pairing in a palindromic sequence. It is likely that such hairpins at the end of RNA promote its dissociation from DNA. Termination can be prevented by an anti-terminator protein, which allows the polymerase to ignore the terminator signal.
A unique distinction between prokaryotic and eukaryotic RNA synthesis is the temporal relationship between its synthesis and utilization in information transfer. Prokary-otic transcription of mRNA is linked to its reading on the ribosome for protein synthesis. Thus, even before transcription is terminated, the 5′ terminal region of the nascent mRNA is complexed with a ribosome for initiation and propagation of protein synthesis. In the case of eukary-otes, transcription occurs in the nucleus, from which the RNA has to be transported to the endoplasmic reticulum with ribosomes in the cytoplasm. Two sequence motifs that are common constituents of promoters in prokary-otic genomes and are nominally referred to as -35 and -10 sequences signify that the midpoint of these sequences are located 35 and 10 bp 5′ of the start site of transcription. However, the exact distance is somewhat variable for different genes. The consensus -35 sequence is TTGACA, and the consensus of -10 is TATAAT. However, both of the sequences are also somewhat variable. The strength of a promoter, i.e., how efficiently it is recognized for transcriptional initiation, depends on the exact sequence of the -35 and -10 sequences and possibly the intervening "spacer" sequences as well. The promoter strength can vary widely among genes, and mutations in the -35 or -10 sequence in a particular gene can dramatically affect its promoter strength.
B. Regulation of Transcription in Bacteria
Unlike replication of the complete genome, which is essential for cellular propagation, not all genes need to be transcribed in a particular cell for its survival. Synthesis of mRNA is required for generation of proteins. Because not all proteins are required at all times for cellular survival and metabolism, both in prokaryotes and eukary-otes, and many proteins are expressed only in specific stages of development and differentiation in higher eu-karyotes, a gene’s transcription is often highly regulated. Furthermore, the stability of mRNAs and the proteins they encode vary over a wide range. Thus, different mRNAs are not made at the same rate. Additionally, the bulk of RNA, and in fact a large fraction of the cell mass, consists of ribosomal and transfer RNAs needed for carrying out protein synthesis. Both ribosomal and transfer RNAs are extremely stable.
Regulation of transcription, first investigated in bacterial viruses, primarily in E. coli, an intestinal microbe and its bacteriophage k, is the foundation of molecular genetics. The ease of generating and manipulating mutants of various genes in E. coli and k led to the discovery of repressors, which are proteins that bind to operator sequences of genes and turn off transcription. The genes that were originally studied encode enzymes for sugar (lactose and galactose) metabolism. Inactivation of these genes and their expression could be studied because the proteins are not essential for bacterial survival. An activator needed for expression of lactose-metabolizing p-galactosidase was identified; it is downregulated in the presence of glucose ("glucose effect") and upregulated by binding to 3′-5′ cyclic AMP.
Significant advances in elucidating the mechanism of transcriptional regulation came from the life cycle studies of the lysogenic k virus, whose virus-specific proteins are not expressed in the lysogenic state, when its duplex DNA genome is linearly integrated in the host chromosome. Here again, both positive and negative regulatory mechanisms are in play to fine tune the expression of genes from a low maintenance level during lysogeny to large-scale expression of the viral genome when the lysogenic virus enters the lytic phase of growth and exploits the host cell synthetic machinery for replication of its own viral DNA, RNA, and proteins.
C. Eukaryotic Transcription
The fundamental process is identical in prokaryotes and eukaryotes, in that an RNA polymerase complex binds to the promoter and initiates transcription at a start site downstream to the promoter. De novo initiation of an RNA chain occurs with a purine nucleotide and creation of a transcription bubble with the open complex. The transcription complex can slide back along the nascent chain and en-donucleolytically cleave off the 3′ segment, then moves forward along the DNA template chain; termination occurs at specific regions in the genes.
In spite of this similarity, however, the details are very different in eukaryotic cells and are summarized as follows.
1. Eukaryotic RNA polymerases contain many more subunits, located in the different regions of the nucleus. Pol I, specific for synthesizing rRNA, is located in the nu-cleolus, a specialized structure within the nucleus, while Pol II and Pol III are in the nucleoplasm. These enzymes have 8-14 subunits with a total molecular mass >500 kD. The large subunits have some sequence similarity with the bacterial RNA polymerases. RNA polymerases of mitochondria and chloroplasts are phylogenetically closer to bacterial RNA polymerase, commensurate with the fact that the target genes of these enzymes are fewer and much smaller in organelles, which are thought to have arisen by symbiotic acquisition of bacteria by primitive eucaryotes.
2. The promoter composition and organization of eu-karyotic polymerases are quite specific for each poly-merase. The promoters of rRNA genes contain a core and an upstream control element which is needed for high promoter activity. Two ancillary factors, UBFl and SLl, bind to these sequences. Although SLl binds only after UBFl in a cooperative fashion, SL1 is a a-factor with four proteins among which TBP is also required for initiation by the other polymerases. Pol I is akin to Pol III in that it utilizes both upstream and downstream promoters. There are two types of internal promoters with distinct sequence boxes. One transcription factor (TFIIIB) is required for initiation of RNA synthesis by Pol III. Other factors (TFIII A and TFIII C) help TFIII B bind to the right location and act as positioning factors for correct localization of Pol III initiation.
Pol II is the most versatile and widely utilized RNA polymerase in vivo and absolutely needs auxiliary, transcription factors (TFII) whose requirement is dependent on the nature of promoters.
3. The nature of eukaryotic promoters is quite different from the prokaryotic promoters. In addition to the bipartite promoter of Pol I, both Pol II and Pol III have a "TATA box" located about 25 bp upstream of the start site in Pol II responsive genes. The 8-bp sequence consists of only A^T base pairs and is surrounded by G^C pair-rich sequences. Interestingly, the TATA box is quite similar to the -10 sequence in E. coli promoters.
There is a second element called a CAAT box, usually about -15 bp 5′ of the TATA box. Alternatively a G^C rich sequence is present in some promoters, often at position -90. The consensus GC box sequence is GGGCGG, of which multiple copies are often present and occur in both orientations. These elements are not all present in all promoters; it appears that they work in a "mix and match" fashion. These boxes, and also a octamer box, bind to specific trans-acting factors and are engaged in multiple protein interactions among themselves as well as with components of the RNA Pol II holoenzyme.
There is no significant homology among transcription start sites of various genes, except for the tendency for the first base in the transcript to be an A flanked on either side by pyrimidines. This region is defined as the initiator.
The first step in transcriptional initiation of a TATA-containing promoter is the binding of the factor TFIID to the region upstream of the TATA site. The TATA-binding protein, TBP, which specifically binds to the TATA box, is a component of the TFIID complex, along with other proteins collectively called TAFs (TBP-associated factors). TAFs can be variable in the TFIID complex, both in species and amounts, and provide the promoter specificity for initiation. Some TAFs are tissue specific. TFIID has a molecular mass of 800 kD, containing 1 TBP and 11 TAFs. TBP acts as a positioning factor and is able to interact with a wide variety of proteins, including Pol II and Pol III. It binds to the minor groove of the DNA double helix and makes contact with other factors which mostly bind to the major groove and can make multiple contacts. By bending the DNA at the binding site, it appears to bring the factors and RNA polymerase into closer proximity.
Although TBP is utilized by both Pol II and Pol III, TFIID is the specific complex for Pol II recognition of a promoter. Other transcription factors (e.g., TFIIA) bind to the TFIID promoter complex and cover increasing segments of DNA. In addition to TFIIA, these include TFIIE, TFIIF, TFIIH, and TFIIJ. Most of the TFII factors are released from the transcription complex before Pol II leaves the promoter and carries out chain elongation. Interestingly, the same general transcription factors, including TFIID, bind to the TATA-less promoter, even though TATA binding by TBP is not available.
There is an important contrast in the assembly of RNA polymerase complexes in eukaryotes and prokaryotes. E. coli RNA polymerase binds to the promoter as a complex with the a-factor, providing the specificity for initiation but not elongation. Eukaryotic Pol II, on the other hand, goes through a much more complex choreography because of the prerequisite for binding to the promoter by other transcription factors. This dichotomy reflects the complex structural organization of the eukaryotic genome and the presence of a much larger number of genes with their complex regulation. Such regulation is not only dependent on the environment, but also on the stage of development and differentiation, at least in the metazoans.
4. A unique difference between prokaryotic and eukaryotic transcription is that in prokaryotes a single mRNA containing many genes can be transcribed from the DNA template as a single transcription unit, coupled with their direct translation on ribosomes into discrete polypeptides. This process reflects the fact that genes which encode enzymes in a given pathway are often clustered in an operon and are co-ordinately regulated.
In contrast to the synthesis of polycistronic mRNA in E. coli and other bacteria, eukaryotic transcription units usually consists of single genes. This characteristic may also reflect uncoupled transcription and translation in these organisms. Thus, heterogeneous nuclear RNA (hnRNA) is synthesized in the nucleus and then transported to the cytoplasm along with its processing into mature mRNA including splicing, addition of poly(A) tail at the 3′ end, and capping at the 5′ end. Subsequently, the RNA is translated on ribosomes (endoplasmic reticulum). Thus, synthesis and utilization of mRNA are temporally and spatially separated.
D. RNA Splicing in Metazoans
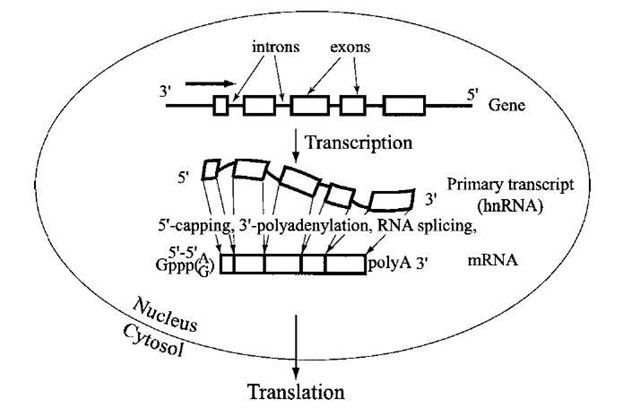
The central dogma of molecular biology that the information flow from DNA to RNA to protein involves colinearity of the sequences of the monomer units is somewhat violated in metazoans because of the presence of interrupted or fragmented genes (Fig. 8). Thus, while the polypep-tide sequence is colinear with the codons of the coding sequence in the mRNA, the RNA itself is not collinear with the gene from which it is transcribed. In other words, the gene contains additional intervening sequences called introns, which are transcribed but whose RNA sequence is subsequently removed from the final mRNA containing the coding sequence. The primary gene transcripts of nuclear genomes, called heterogeneous nuclear RNA (hnRNAs), are present in a form of protein-bound particles (ribonucleoprotein particles, or hnRNP). RNA splicing is the process of excising introns from hnRNAs, and contiguous exons are then joined to form mature mRNAs, which are subsequently translocated to cytoplasm and are used as templates for translation (Fig. 8). The cleavage and rejoining occur at specific junctions between exons and introns, so that there are no errors in mature mRNA. First, two adjacent exons are aligned, while the intervening intron is extruded, forming a loop ("lariat") structure. Then the upstream exon is cleaved and joined to the downstream exon via a transesterification reaction. In most cases, two factors are essential for this process. One, the cis-elements in introns and exons, is the signaling sequences for the exact junction sites. The other is the splicing machinery, consisting of several small ribonucleoprotein particles (snRNP; U1, U2, and U4-U6), each of which contains small RNA molecules and proteins. The U1 and U2 snRNPs contain RNA complementary to the intron cis-element and catalyze the formation of the intron lariat, while two adjacent exons are aligned together. With other snRNPs forming an intermediate complex (spliceosome), U6 catalyzes the transesterfication. It should be noted that introns in RNA of some lower eukaryotic species are autospliced and therefore do not require snRNPs.
FIGURE 8 A schematic representation of RNA splicing. The coding sequence in metazoan genomes is usually present in segments (exons; indicated by boxes) interspersed between noncod-ing introns. After synthesis of the primary RNA transcript (called heterogeneous nuclear RNA or hnRNA), the intron sequences are removed by precise cleavage and rejoining is mediated by the spliceosome complex, so that the resulting mature mRNA contains a correctly juxtaposed coding sequence for the polypeptide. The mRNA is also "capped" by 5′-5′ linkage with GMP, and a tail of poly(A) is added at the 3′ terminus to increase the stability of mRNA and to enhance its efficiency in directing protein synthesis when the mRNA is transported from the nucleus to the cytoplasm.
Termination of eukaryotic transcription is coupled with processing. The mature rRNA is obtained by cleavage of a larger primary transcript synthesized by Pol I. Termination of Pol II transcription occurs at a repeat sequence of U, as in the case of E. coli RNA polymerase, but without the presence of a hairpin structure. More importantly, the 3′ termini of mRNAs are generated by cleavage of primary precursor transcripts followed by addition of a tail of poly(A), a homopolymer of up to several hundred AMP residues synthesized by poly(A) polymerase in a template-independent reaction.
E. Regulation of Transcription in Eukaryotes
While both prokaryotic and eukaryotic genes are regulated by activators and repressors, enhancer elements are unique to eukaryotic genes and can profoundly increase the rate of transcription. These elements are located at a variable distance from the basic promoter itself, can be present both upstream or downstream to the promoter, and, in fact, can even be within the transcription unit. One unexpected feature is that they can function in either orientation and can activate any promoter located in the vicinity.
Upstream activating sequences (UAS) have been identified in yeast and are analogous to enhancers in the mammalian genes. Based on the known properties of enhancers, it appears that the presence of these sequences affects chromatin structure and/or the helical structure of the DNA template itself. Further studies are needed to test other possibilities as well, e.g., whether the enhancer provides an entry point for the transcription complex or is needed to place the template at the nuclear matrix where transcription takes place.
Positive and negative regulation of prokaryotic genes is achieved by binding of activators and repressors, respectively, to their cognate binding sites in the genes. Down-regulation is more common, at least in E. coli, than positive regulation. In fact, the same protein can provide dual functions in a few cases, depending on the location of the sequence motif.
In contrast, because of the complexity of chromatin structure and genomic organization development, differentiation, and cell cycle-specific synthesis of proteins, regulation of eukaryotic genes is extremely complex. This is evident from the large number of families of regulatory trans-acting factors which recognize similar if not identical sequence motifs in different genes. Sometimes, these factors have a distinct modular structure—one module for binding to target DNA sequence and another for interaction with components of the transcription apparatus.
On top of these complexities, the signal for initiation of transcription may be extracellular, e.g., a growth factor which induces cell proliferation. A highly complex signaling cascade is initiated in response to the first signal. The external ligand first binds to its receptor on the cell surface, followed by internalization of the receptor ligand complex. A series of reversible chemical modification (mostly phosphorylation of the regulatory proteins) finally activates the ultimate transcription factors, which then trigger transcription of target genes.
The unique difference between the eukaryotes and prokaryotes is in the utilization of transcription factors. In bacteria, one factor is usually specific for one gene or one regulatory unit. In eukaryotes, on the other hand, a single factor activates multiple target genes.
Prokaryotic regulatory processes have been elucidated in remarkable detail by utilizing the power of molecular genetics, including "reverse genetics" by which the chromosomal genes in the organism could be mutated at specific sites and the mutant gene products purified and characterized. Furthermore, these genes can be expressed in the episomal state by introducing them into autonomously replicating recombinant plasmids.
Commensurate with the significantly higher complexity and size of the genome and differentiation and developmental stages in metazoans, gene regulation in these organisms is very complex and occurs at many levels. Sets of genes are activated at distinct stages of differentiation and development of multicellular organisms in order to encode proteins which are required for specialized functions of the cells in these stages. In contrast, certain "housekeeping" proteins, including enzymes for metabolism and synthesis of all cellular components (i.e., RNA, DNA, structural proteins, and lipids), as well as enzymes for biosynthetic and degradative pathways, are needed in all cell types and developmental stages. Most somatic cells in adult mammals are nondividing and therefore do not require DNA synthesis machinery. However, all cells require transcription for generating proteins for other cellular functions. Unraveling the molecular mechanisms of regulation is the major focus of current research in molecular biology. The regulatory process is affected by multiple parameters.
Many genes are activated due to external stimuli, e.g., exposure to hormones and growth factors. In these cases the extracellular signal often acts as a ligand to bind to cell surface receptors which activate the trans-acting factor(s) via multiple steps of signal transduction.
1. Regulation of Transcription via Chromatin Structure Modulation in Eukaryotes
The eukaryotic genome is organized at multiple levels, starting with the nucleosome core as described earlier. The nucleosomes are organized in a higher order chromatin structure due to increasing compaction of DNA: from 2-nm-wide naked DNA fiber to metaphase chromosomes of microscopic width. The DNA template has to be accessible to transcription machinery containing RNA poly-merase; transcriptionally inactive, highly compacted chro-matin maintains its structure by multiple protein-protein and protein-DNA interactions, which are yet to be elucidated. However, it is now clear that at the nucleosome level, it is the strength of interaction between histones and DNA which regulates accessibility of the DNA to the transcription machinery, a process controlled by acetyla-tion and phosphorylation of core histones. Multiple his-tone acetylases and deacetylases, which are themselves regulated, modulate chromatin structure. As stated previously, large protein complexes named SWI and SNF modulate chromatin structure in an energy-dependent process which may be responsible for the differentiation/ development-dependent turning on or off of specific sets of genes.
2. CpG Methylation-Dependent Negative Regulation of Genes
In addition to histone modification, DNA itself was found to be modified, most commonly by methylation at the C-5 position of cytosine, but only when it is present as a CpG dinucleotide. Such methylation, catalyzed by specific methyltransferases, invariably inhibits gene expression, which was unequivocally established in the genomes during embryonic development. Sets of genes are selectively methylated or demethylated in the CpG sequences, most commonly in the genes’ promoter regions, leading to their activation or repression. Proteins that bind to methylated CpG sequences have been implicated in the control of histone deacetylation, thereby leading to closing of the promoter.
F. Fidelity of Transcription (RNA Editing)
The informational content of gene transcripts can be altered during or after transcription by a process collectively called RNA editing. The information changes are carried out at the level of mRNA. RNA editing appears to be a widespread phenomenon for both normal and aberrant RNA processing in organelles and nuclei. It was first discovered in the mitochondria of kinetoplasts in protozoa. Two types of RNA editing have been observed: (1) alteration of coding sequence by nucleotide insertion and/ or deletion and (2) base substitution. In mammalian cells, editing of an individual base in mRNA can cause a change in the sequence of the protein. Such changes can occur by enzymatic deamination in which C is converted to U or A is converted to hypoxanthine. Change of U to C has also been observed in many plants. The (mitochondrial) mRNAs of several kinetoplastid species (Crithidia, Trypanosoma, etc.) were found to be edited by the insertion and deletion of U’s at many sites in mRNAs. The editing process uses a template consisting of a guide RNA (gRNA) whose genes function as independent transcription units. The gRNAs are generally 55-70 nucleotides in length and complementary to the mRNA for a significant distance including and surrounding the edited region. The gRNA dictates the specificity of uridine insertions by its pairing with the pre-edited RNA, but also provides the U residues that are inserted into the target RNA by transesterification reactions; the reaction proceeds along the pre-edited RNA in the 3′-5′ direction. The RNA editing process reveals the existence of a previously unrecognized level for the control of gene expression. Recognition of this process has resulted in an expansion of the central dogma. Multiple RNA editing processes play a significant role in normal physiological processes, as well as being responsible for some disease.