2. Genomes
The majority of plant viruses (71%) have (+)-strand (messenger-sense) RNA as their genome; genomes are also found that consist of double-stranded DNA (4%), single-stranded DNA (9.5%), double-stranded RNA (5%), and (-)-strand RNA (10.5%). The double-stranded DNA genome plant viruses replicate by reverse transcription, and there are no plant viruses known with double-stranded DNA genomes that replicate the DNA directly.
2.1. Genome, Structure, Organization, and Expression
Plant viruses encode products that facilitate their replication, encapsidation, and in most cases, their movement within and between plants (see Virus Infection, Plant). Expression of these products is constrained by the limitations of eukaryotic ribosomes being able to translate only the 5′ open reading frame of a messenger RNA (mRNA). Both plant and animal viruses, have developed various strategies for overcoming this limitation. The genome organizations of typical members of the 48 genera (where sequence data are available) are illustrated in Figure 1.
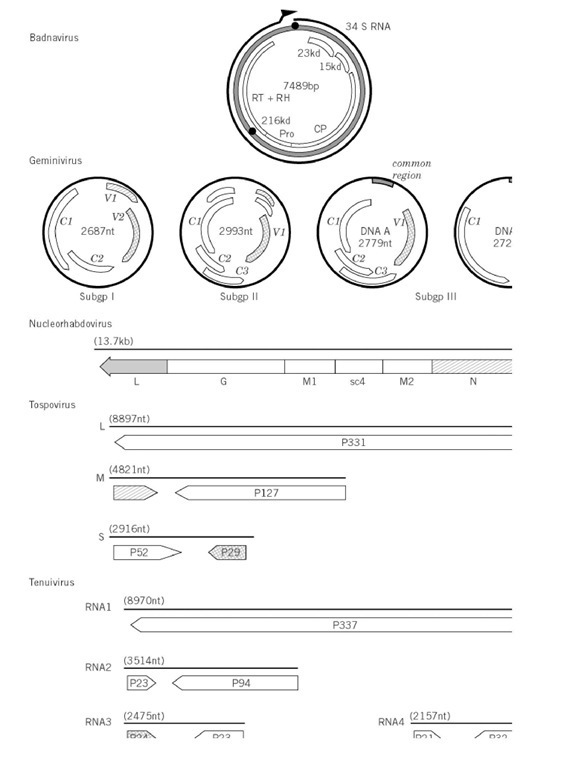
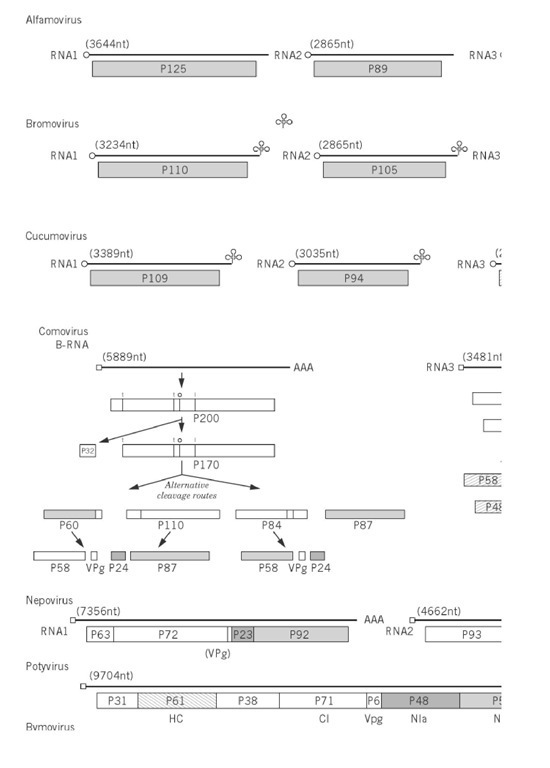
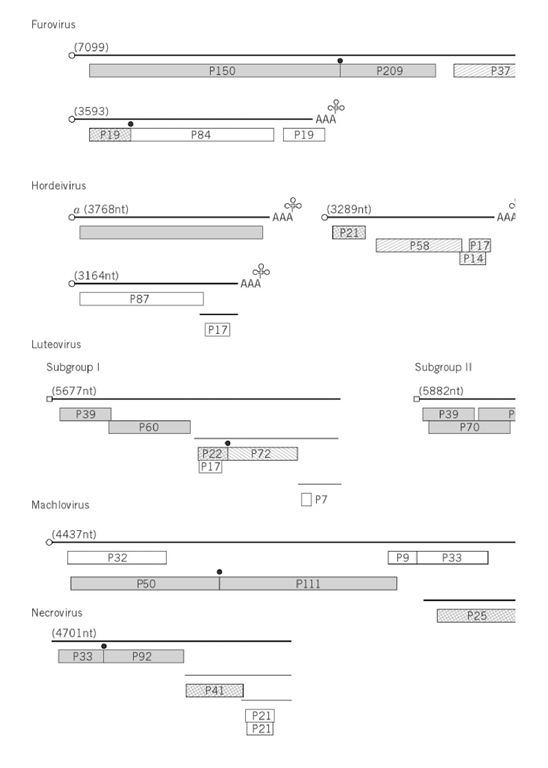
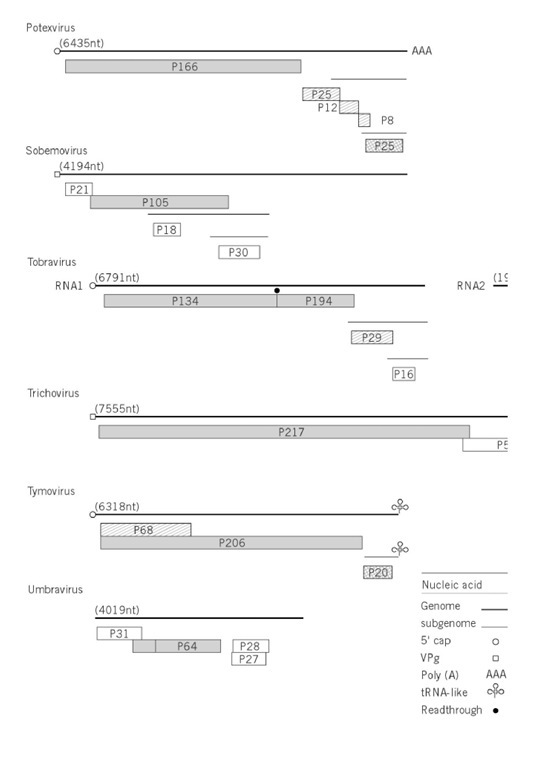
Figure 1. Genome organizations of groups of plant viruses. The illustration is of the type member of the group (Table 1 ) except subgroup III geminiviruses, African cassava mosaic virus; nucleorhabdovirus, sonchus yellow net virus; nepovirus, tomato blackring virus; carlavirus, potato virus M; dianthovirus, red clover necrotic mottle virus; umbravirus, groundnut rosette virus. The genome is represented by two concentric circles for double-stranded circular genomes; one circle for single-stranded circular genomes; a straight line for linear genomes; subgenomic mRNA are shown as thin lines. The size of the genome is given in nucleotides (nt or base pairs, bp). The terminal features of the genomes, when they are known, are listed on the figure. The gene products are indicated by boxes, with the directions of those of ambisense genomes given by an arrow. The molecular size of the gene product is x10-3. The symbols for functions of gene products and expression by read through of a weak stop codon are indicated on the figure.
Figure 1. Genome organizations of groups of plant viruses. The illustration is of the type member of the group (Table 1 ) except subgroup III geminiviruses, African cassava mosaic virus; nucleorhabdovirus, sonchus yellow net virus; nepovirus, tomato blackring virus; carlavirus, potato virus M; dianthovirus, red clover necrotic mottle virus; umbravirus, groundnut rosette virus. The genome is represented by two concentric circles for double-stranded circular genomes; one circle for single-stranded circular genomes; a straight line for linear genomes; subgenomic mRNA are shown as thin lines. The size of the genome is given in nucleotides (nt or base pairs, bp). The terminal features of the genomes, when they are known, are listed on the figure. The gene products are indicated by boxes, with the directions of those of ambisense genomes given by an arrow. The molecular size of the gene product is x10-3. The symbols for functions of gene products and expression by read through of a weak stop codon are indicated on the figure.
Figure 1.Genome organizations of groups of plant viruses. The illustration is of the type member of the group (Table 1 ) except subgroup III geminiviruses, African cassava mosaic virus; nucleorhabdovirus, sonchus yellow net virus; nepovirus, tomato blackring virus; carlavirus, potato virus M; dianthovirus, red clover necrotic mottle virus; umbravirus, groundnut rosette virus. The genome is represented by two concentric circles for double-stranded circular genomes; one circle for single-stranded circular genomes; a straight line for linear genomes; subgenomic mRNA are shown as thin lines. The size of the genome is given in nucleotides (nt or base pairs, bp). The terminal features of the genomes, when they are known, are listed on the figure. The gene products are indicated by boxes, with the directions of those of ambisense genomes given by an arrow. The molecular size of the gene product is x10-3. The symbols for functions of gene products and expression by read through of a weak stop codon are indicated on the figure.
Figure 1.Genome organizations of groups of plant viruses. The illustration is of the type member of the group (Table 1 ) except subgroup III geminiviruses, African cassava mosaic virus; nucleorhabdovirus, sonchus yellow net virus; nepovirus, tomato blackring virus; carlavirus, potato virus M; dianthovirus, red clover necrotic mottle virus; umbravirus, groundnut rosette virus. The genome is represented by two concentric circles for double-stranded circular genomes; one circle for single-stranded circular genomes; a straight line for linear genomes; subgenomic mRNA are shown as thin lines. The size of the genome is given in nucleotides (nt or base pairs, bp). The terminal features of the genomes, when they are known, are listed on the figure. The gene products are indicated by boxes, with the directions of those of ambisense genomes given by an arrow. The molecular size of the gene product is x10-3. The symbols for functions of gene products and expression by read through of a weak stop codon are indicated on the figure.
Figure 1.Genome organizations of groups of plant viruses. The illustration is of the type member of the group (Table 1 ) except subgroup III geminiviruses, African cassava mosaic virus; nucleorhabdovirus, sonchus yellow net virus; nepovirus, tomato blackring virus; carlavirus, potato virus M; dianthovirus, red clover necrotic mottle virus; umbravirus, groundnut rosette virus. The genome is represented by two concentric circles for double-stranded circular genomes; one circle for single-stranded circular genomes; a straight line for linear genomes; subgenomic mRNA are shown as thin lines. The size of the genome is given in nucleotides (nt or base pairs, bp). The terminal features of the genomes, when they are known, are listed on the figure. The gene products are indicated by boxes, with the directions of those of ambisense genomes given by an arrow. The molecular size of the gene product is x10-3. The symbols for functions of gene products and expression by read through of a weak stop codon are indicated on the figure.
2.2. Double-stranded DNA Viruses
The genomes of the two genera of double-stranded DNA viruses are similar in that they are circular molecules of about 7.2 to 8.0 kbp, with one or more discontinuity in each strand; these discontinuities are described in more detail in Cauliflower mosaic virus. The viral DNA of both these genera is transcribed asymmetrically to give a more-than-genome length RNA. The genomes of the badnaviruses encode three or four proteins. The largest badnavirus open reading frame is expressed as a polyprotein (Fig. 1), which is processed to yield the viral coat protein (CP), a carboxyl proteinase (Pro), and reverse transcriptase plus ribonuclease H (RT + RH) (the enzymes involved in reverse transcription). The three open reading frames of most badnaviruses are expressed from the genome-length RNA transcript by unusual mechanisms involving ‘ribosomal shunting’ and leaky translation initiation (see (5) for a review).
2.3. Single-stranded DNA Viruses
Geminiviruses have circular single-stranded DNA molecules of about 2.7 to 3.0 kb. The three genera of the Geminiviridae have similarities in their genome organizations and expression, but there are differences. Subgroups I and II have monopartite genomes, with members of subgroup 1 infecting monocotyledenous plants and those of subgroup II dicotyledons. The genome of most subgroup III members (which infect dicotyledenous plants) is divided between two DNA molecules. Members of subgroups I and II are transmitted by leafhoppers, those of subgroup III by whiteflies. In all cases, the genomes are transcribed bilaterally from a characteristic region near the mapping zero point (termed the common region for subgroup III members), the transcripts terminating on the opposite side of the genome. Thus, one transcript is from the virus-sense DNA and the other from the complementary-sense DNA. Each of these transcripts expresses one or more proteins (Fig. 1) through mechanisms that involve both different transcript origins and splicing. Molecular aspects of geminiviruses are reviewed in (6, 7).
2.4. Double-stranded RNA Viruses
The genome organizations of plant viruses with double-stranded RNA genomes in many cases resemble those of animal-infecting reoviruses. Members of the plant Reoviridae are insect-transmitted, and the virus multiplies in the vector. The genomes of these viruses are divided between 10 or 12 segments of about 2.5 to 0.8 kb that, in most cases, are monocistronic (8). These encode the various proteins that make up the virion structure, together with various nonstructural proteins, including the viral polymerase.
The cryptic viruses (Partitiviridae) have no known vectors and are spread by the vegetative propagation of their hosts. Their genomes have only two monocistronic genome segments, one encoding the viral polymerase and the other the coat protein (9).
Each genomic RNA of these double-stranded RNA viruses has a conserved terminal oligonucleotide sequence that is thought to be genus-specific (8).
2.5. Single-stranded (-)-Sense RNA Genomes
Two of the three families of plant viruses with (-)-sense RNA genomes resemble viruses that infect animals, and they have many common genome organizational features. They also replicate in their insect vectors. The best characterized rhabdovirus, Sonchus yellow net virus, belongs to the Nucleorhabdovirus genus and has a genome organization identical to animal rhabdoviruses, except that it has an extra gene product (sc4) (Fig. 1) (10). The genome length (-)-sense RNA is transcribed into monocistronic subgenomic mRNA, molecules.
The Tospoviruses resemble the animal Phlebovirus genus of the Bunyavirus family in having three genome segments, the largest (L RNA, Fig. 1) of which is (-) sense and the other two having an ambisense expression strategy. In this, the medium and smallest RNA segments of Tospoviruses (M and S RNA) each encode one protein from the 5′ end of the viral-sense RNA and another protein from the 5′ end of the complementary RNA strand (ie, the open reading frame is at the 3′ end of the viral-sense RNA). The ambisense strategy is taken even further in the Tenuiviruses, which have their largest genome segment (RNA1, Fig. 1) as (-)-sense RNA and the other three (RNAs 2, 3, and 4) with the ambisense arrangement. In both the Tospoviruses and Tenuiviruses, the largest RNA encodes the RNA-dependent RNA polymerase. Molecular aspects of tospoviruses are reviewed in (11) and those of tenuiviruses in (12).
2.6. Single-stranded (+)-Sense RNA Genomes
This genome, the most common type found in plant viruses, acts as an mRNA. A range of structures are found at the ends of these RNA (indicated in Fig. 1). Many viruses have a methylated blocked 5 ‘ teminal group
where X(m) and Y (m) are methylated bases. Others have a virus-encoded protein covalently linked to the 5 ‘ end of the genomic RNA. The linkage of this protein, the Vpg, has been identified in some cases and, for cowpea mosaic, has been shown to be a phosphodiester bond between the b-hydroxyl group of a serine residue at the N-terminus of the Vpg and the 5′-terminal uridine residue of the two genomic RNA. At the 3′ end, some viruses have a polyadenylate sequence resembling that of many eukaryotic mRNAs. Other viruses have 3′ sequences that fold to resemble transfer RNA (tRNA). These structures will accept amino acids; eg, that of TMV accepts histidine, BMV and CMV accept tyrosine, and turnip yellow mosaic tymovirus accepts valine. Barley stripe mosaic hordeivirus has a 3′ -terminal tRNA-like structure (accepting tyrosine) and an internal poly(A) tract (Fig. 1). The functions of these 3′ structures are unknown but thought to be involved with replication.
The 5′ noncoding regions of several plant viruses have been shown to have translational enhancing activity. That of TMV, the W sequence, is discussed in Tobacco mosaic virus. The 3′ noncoding regions of some viruses fold as pseudoknots with unknown function.
An apparently wide range of genome organizations and expression is found in viruses with (+)-sense RNA genomes (Fig. 1), but, if we take into consideration the limitations of eukaryotic ribosome translation, certain basic features emerge. There are two predominant methods by which the four or more gene products are expressed from these viral genomes. In the first, exemplified by carmoviruses, luteoviruses, potexviruses (see Fig. 1), and TMV, the 5 ‘ cistron is expressed from the virion RNA, and most of the downstream cistrons are expressed from subgenomic RNAs that are transcribed from the (-)-strand replication intermediate. There are some subtleties in the expression of the 5′ genetic information in that, in eg, carmoviruses and TMV, the first two or three open reading frames are separated by weak stop codons that are read through. A variant of this strategy is to frameshift between the first and second open reading frame of, eg, luteoviruses, when ribosomes change reading frame from one cistron to that of an overlapping cistron. In both read through and frame shift, the larger product is formed to a much lesser extent than the 5 product. The division of the genome into monocistronic units is taken even further in many genera of plant viruses that have multipartite or divided genomes. The genome is divided into two or three segments that are encapsidated separately. For example, members of the Bromoviridae (Alfamovirus, Bromovirus, and Cucumovirus) have their genomes divided into three segments. The larger two segments are monocistronic, whereas the smallest segment is bicistronic but expresses the 3′ cistron from a subgenomic RNA.
The second predominant method by which viral gene products are expressed is by having a single open reading frame on the genomic RNA that is translated to give a polyprotein. This is processed to the functional proteins by one or more proteinases encoded within the polyprotein. Potyviruses, Sequiviruses, and Waikaviruses are examples of viruses with a single genome segment that follow this strategy. Comoviruses, Nepoviruses, and Bymoviruses have their genomes divided into two segments, each of which encodes a polyprotein. The processing pathways can be complex, as shown by Comoviruses (Fig. 1).
2.7. Genome Replication
The replication of the various type of genome is discussed in Virus infection, plant.

![tmp24-111_thumb[2] tmp24-111_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp24111_thumb2_thumb.jpg)
