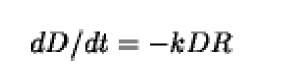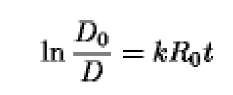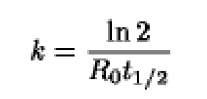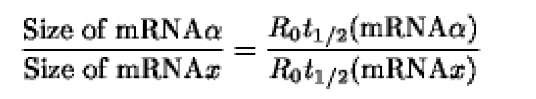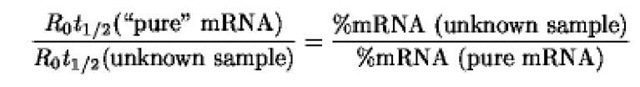1. DNA-RNA Hybridization
During purification of messenger RNA, its quantity and the extent of purification can be determined by hybridization techniques. This also permits determining the size of the mRNA independently by gel electrophoresis.
Hybridization involves annealing a large excess of RNA with double-stranded DNA previously radiolabeled by nick translation or during its synthesis. Such labeling techniques result in a high specific radioactivity, so the necessary excess of RNA can be obtained easily. Heating double-stranded DNA denatures it to its single-stranded components and slow cooling of these single strands permit renaturation. This technique is used in CQt curves to characterize DNA samples. Hall and Spiegelman (1) demonstrated that the same experiment can be performed with heat-denatured DNA and the corresponding mRNA, and slow cooling produces DNA-mRNA hybrids that were observed in cesium chloride density gradient centrifugation. When mRNA was heated with genetically unrelated DNA, no hybrid molecules were formed.
1.1. Kinetics of Association
DNA-RNA hybridization is a second-order reaction (see Kinetics). If t is the time, D the concentration of single-stranded DNA in moles of nucleotides/liter, R the concentration of excess mRNA in the same units, and k the rate constant for the reaction, then the equation describing the reaction is the following:
The value of k is expressed in units of Ms
DNA self-reassociation is avoided by using a low concentration with RNA in excess. Then the RNA hybridizes much faster than the DNA, and its concentration remains practically constant throughout the reaction. The reaction follows pseudo-first order kinetics, and the equation above simplifies to the following:
Rq is the mRNA concentration at t = 0.
By substituting D = A>at t = 0, the integrated equation becomes
from which the value of k can be derived by
When 50% of the DNA (cDNA) strands have been converted into DNA-RNA hybrids, at – ”V-/J becomes D0/2, and R01 becomes Rqt j / 2. This yields
Therefore the velocity of the association reaction is inversely proportional to the value of R0^ / 2, which itself is directly related to the size of a pure RNA or DNA molecule.
1.2. Determining mRNA size and evaluating a given mRNA in an RNA mixture
Knowing the size a of a given mRNA, the unknown size x of another mRNA is determined by the following equation:
For ovalbumin mRNA, which is 2000 nucleotides long, the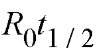 value is
value is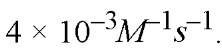 For prolactin mRNA (900 nucleotides long) the
For prolactin mRNA (900 nucleotides long) the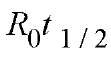 value is accordingly smaller:
value is accordingly smaller:![]() (2).
(2).
The values of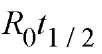 allow determining the fraction of a certain mRNA in a mixture by comparing hybridization of the corresponding cDNA with the corresponding "pure" mRNA and with the same mRNA in a mRNA mixture. Because the size of the desired mRNA is constant, its reassociation rate depends only on its concentration in the mixture, according to the equation
allow determining the fraction of a certain mRNA in a mixture by comparing hybridization of the corresponding cDNA with the corresponding "pure" mRNA and with the same mRNA in a mRNA mixture. Because the size of the desired mRNA is constant, its reassociation rate depends only on its concentration in the mixture, according to the equation
The course of hybridization is monitored by determining the number of DNA-RNA hybrids at various reaction times. This is done in two ways (2):
1. by applying the reaction mixture to a chromatography column of hydroxyapatite in 0.05 M phosphate buffer at pH 6.8 and at a temperature of 60°C. The single-stranded DNA elutes with 0.14 M phosphate buffer, whereas the double-stranded DNA-RNA molecules elutes with 0.45 M phosphate buffer.
2. by digestion with S1 nuclease, which digests single-stranded DNA, whereas RNA-DNA hybrids remain unaffected and are precipitated with trichloroacetic acid.
2. Determining the Number of Different Sequences in mRNA Transcribed from a cDNA Library
To judge whether a cDNA library is representative, ie, whether the number of different clones carrying a particular cDNA insert reflects the relative fraction of the corresponding mRNA molecules in the cell from which the library has been prepared, the number of different mRNA sequences in the mRNA population is determined. This is done by hybridizing highly labeled cDNA with an excess of mRNA. The amount of hybridized DNA at any time t is plotted as a function of R0.
The kinetic curve yields the number of kinetic classes of mRNA molecules and their R0^ / 2 values.
The values are corrected to obtain the values that would have been obtained if each of the classes had been considered on its own. The values are also corrected for the nonspecific poly-dA sequences in the cDNA from the mRNA poly A tails, which do not contribute to hybridization. Then the corrected R>t1 / 2 values are used to determine the number of different sequences in each kinetic class by comparing them with the values of mRNA of known complexity ((3),(4)).