The dversity of function of RNA molecules in living organisms ranges from the enzyme-like activity of ribozymes to storage of genetic information in RNA viruses. RNA molecules adopt diverse structures in response to these functional requirements. RNA has a covalent structure very similar to that of DNA, the only differences being the change from a 2 ‘ -deoxyribose sugar in DNA to a ribose sugar in RNA and from a methyl group in thymidine to a hydrogen atom in uracil. However, their functional differences lead to correspondingly different structures. The requirement for storage of genetic information imposes the double-helical structure on most DNA molecules, whereas RNA molecules adopt an array of structures to rival protein structures in their complexity. This is not the result of an intrinsic limitation of DNA stereochemistry, but rather the result of different functional requirements.
It is convenient to describe RNA structure in hierarchical terms (Fig. 1), comparable to those used in describing protein structure: primary, secondary, tertiary, and quaternary structures. The primary structure refers to the sequence of an RNA molecule. Unlike proteins, which in most cases function only when properly folded, many RNAs function as unstructured, single-stranded species. For example, messenger RNA must be unfolded for the genetic message to be translated, and stable RNA secondary structures inhibit protein biosynthesis.
Figure 1. Hierarchy of RNA folding. (a) The primary structure corresponds to the RNA sequence. (b, and c) The secondary structure (in this case, two stem-loops or hairpins) form by Watson-Crick base pairing between complementary nucleotides. (d) Tertiary interactions (coaxial stacking of double helices) lead to the final three-dimensional structure (in this case, a pseudoknot).
1. Secondary Structure of RNA
Secondary structure in DNA and RNA is dominated by Watson-Crick base pairing, leading to the formation of double-helical structures of varying length. Isolated base pairs are not thermodynamically stable, but formation of several consecutive base pairs readily occurs, resulting in a variety of possible arrangements (Fig. 2). These so-called secondary structure motifs represent the building blocks through which the most complex RNA three-dimensional structures are constructed. There is a fundamental difference between RNA and protein structures. Protein secondary structure is generally only marginally stable in the absence of stabilizing tertiary structure interactions, whereas RNA secondary structure is often stable on its own. Thus, formation of the secondary structure dominates the process of RNA folding, and RNA tertiary structure forms through relatively weak interactions between pre-formed secondary structure motives. Because of this property, RNA secondary structure can often be predicted successfully from thermodynamics
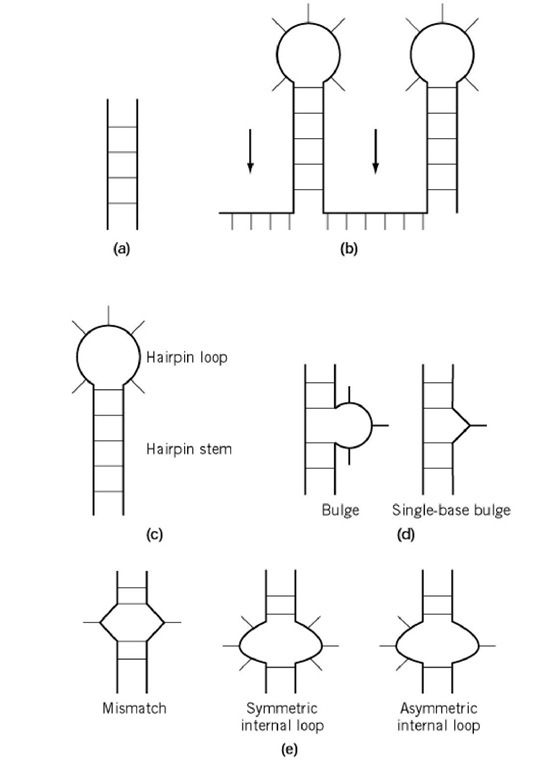
Figure 2. RNA secondary structure motifs. (a) Duplexes; (b) Single-stranded regions; (c) hairpins; (d) bulges; (e) internal loops.
RNA secondary structure is dominated by the formation of double helices stabilized by Watson-Crick base pairs between complementary stretches. Unlike DNA, these helices are relatively short, seldom more than 8 to 10 base pairs in length, and are interrupted by single-stranded nucleotides (1) or phylogeny (2). forming loop elements: hairpins, bulges, and internal loops (Fig. 2). These, together with the helical junction that is formed when more than two double helices come together, are the secondary structure motives and the building blocks upon which the most complex RNA structures are built.
In many RNAs, more than half of all nucleotides are incorporated into double-stranded helices. Duplex regions can form through very long-range interactions, and these interactions are crucial to determine and stabilize the overall fold of an RNA molecule. For example, opposite strands in double helices within ribosomal RNA can be separated by as many as 2000 nucleotides. In RNA, G-U pairs are almost as common as the canonical G-C or A-U base pairs, and they introduce slight distortions in double-helical structure that are recognized by proteins and other RNAs (3, 4). RNA (and DNA) double helices have an antiparallel right-handed helical conformation. RNA double helices adopt the A-form structure, which differs significantly from the canonical B-form adopted by DNA double helices (see DNA Structure). RNA duplex structures are not uniform, although their variability is less than that in DNA. These variations depend on the sequence and structural context of the helix relative to the global three-dimensional structure. A-form double helices differ from B-DNA in the conformation of the sugar and the displacement of the bases from the helical axis. These local differences lead to very diverse shapes, with profound consequences for recognition by proteins and other ligands (5).
The most common element of RNA secondary structure is the hairpin (or stem-loop) (Fig. 2). For example, bacterial 16S ribosomal RNAs contains approximately 30 phylogenetically conserved hairpins (2). A hairpin forms when the phosphodiester backbone folds back on itself to form a double-helical tract (called the stem), leaving unpaired nucleotides to form a single-stranded region, called the loop. Hairpin loops represent the most extensively studied RNA structure motif (apart from double helices). Small hairpin loops contain a high degree of structure, whereas longer loops (containing more than 7 to 8 unpaired nucleotides, such as those present in transfer RNA) are generally more poorly structured and thermodynamically less stable. Most loops in ribosomal RNA are small, 4 to 9 nucleotides in length, perhaps reflecting these thermodynamic preferences.
1.1. Tetraloops
Hairpins containing four nucleotides (tetraloops) are unusually common in cellular RNAs, and their sequences cluster within three exceptionally common families: UNCG, GNRA, and CUUG (where N is any of four nucleotides and R is a purine). For example, 70% of all tetraloops in 16S-like ribosomal RNAs from all organisms belong to the UNCG and GNRA families. The exceptionally common tetraloop structures have high thermodynamic stabilities, UNCG being the most stable. Capping a double-helical tract with a UNCG tetraloop is thermodynamically equivalent to extending the double-helical stem by two base pairs. Despite the differences in sequence, there are extensive structural similarities between these three families. In each of them, the first and last unpaired nucleotides form non-Watson-Crick base pairs to close the loop, reducing the number of unpaired nucleotides to only two. Longer loops, such as those found in tRNA, preserve extensive base stacking interactions that presumably stabilize the loop structure, but are generally characterized by significant conformational flexibility.
1.2. Bulges and Internal Loops
Bulges and internal loops form when two double-helical tracts are separated on either one (bulge) or both strands (internal loops) by one or more unpaired nucleotides. Internal loops containing equal numbers of bases on each strand are symmetric, whereas they are asymmetric when the number of bases are different. For example, single base mismatches are symmetric internal loops of two nucleotides. The presence of an internal loop or bulge reduces the thermodynamic stability, when compared to a perfect double helix, but unpaired nucleotides are more readily accessible to protein or nucleic acid ligands, which often recognize such sites. Non-Watson-Crick base pairs readily form within internal loops, while unpaired nucleotides within a bulge may stack within the helix or be bulged outside. The presence of an internal loop or bulge can induce bending in an RNA molecule; the extent of bending depends on the RNA sequence within the loop and can change upon ligand binding (6, 7). Thus, these motifs are ideal sites for conformational switches, where ligand binding can result in long-range conformational changes. 2. Tertiary Structure of RNA
Interactions between two or more secondary structure elements give rise to RNA tertiary structure and define the overall folding of RNA molecules. In essentially all cases investigated thus far, RNA secondary structure elements maintain their three-dimensional structure even when extracted from very complex tertiary structures. Noncanonical base pairs, unpaired bases, and the backbone functional groups (the negatively charged phosphate groups and the unique 2 -hydroxyl group of RNA) are very important for tertiary interactions. Unpaired bases can twist or flip out of an helical patch to define unique surfaces for recognition by other RNAs during formation of the tertiary structure. The RNA secondary structure helps in orienting key residues into appropriate positions for tertiary interactions to occur. For example, the geometry of the four-way junction in tRNA, combined with a conserved length of the double-helical regions, help in positioning T-loop and D-loop nucleotides in close proximity to facilitate the tertiary loop-loop base pairs that define the L shape of all tRNAs (8).
Tertiary interactions often consist of base stacking and hydrogen bond interactions. Helical stacking between the terminal base pairs of two helices enables the building of the molecule into an extended helix (coaxial stacking) (Fig. 1). A common example of a structural module built upon base-stacking interactions is provided by the adenosine platform motif . Consecutive adenine bases within an internal loop can form a pseudo-A-A non-Watson-Crick base pair, to create a platform capable of mediating long-range tertiary interactions (9). Base triplets form when a preformed base pair becomes involved in another set of hydrogen bonds with a third base (8); this can occur on either the minor groove or major groove side of an RNA double helix. Hydrogen bonds between bases and backbones are observed when double helices are packed together in compact structures (10). Finally, divalent metal ions, especially hydrated magnesium ions, are often used to screen the negatively charged phosphate groups along the helical backbone, in order to build a compactly folded structure with close packing of negatively charged phosphates (11).
Unpaired nucleotides embedded within a secondary structure motif often form tertiary structures by interacting with unpaired nucleotides from another secondary structure module. These interactions can involve any secondary structure motif: hairpin loops, internal loops, and bulge loops. The nature of the tertiary contacts can be intercalation, base triplet formation, and Watson-Crick base pairing between complementary loop sequences. The loop-loop interaction module is best illustrated by the tertiary interaction of tRNA (8). The folding of tRNA is initiated by the formation of four hairpin loops via Watson-Crick base pairing, resulting in the organization of a four-way helical junction. Tertiary interactions, by helical stacking, further organize the tRNA into two extended helical domains. This arrangement positions all unpaired residues in the loop regions close in space and facilitates the formation of the tertiary base pairing and base triplets that lock the tRNA into the L-shaped three-dimensional structure.
2.1. Pseudoknots
Pseudoknots form when complementary primary sequences of a hairpin or internal loop and a single-stranded region interact with each other by Watson-Crick base pairing (12). When a pseudoknot forms between a hairpin loop and a complementary single-stranded region, then formation of two alternative hairpin structures can occur (Fig. 1). The formation of a pseudoknot creates an extended helical region through helical stacking of the hairpin double-helical stem and the newly formed loop-loop interaction helix. Although the pseudoknot is only marginally more stable than the two hairpins, tertiary interactions (such as base triplets) between unpaired nucleotides in the bridging loops and between base pairs within the extended helix can increase the stability of this structure.
2.2. Magnesium Ions
Double-helical regions must be packed together to build compact RNA structures, as found for example in the catalytic core of catalytic RNA ribozymes. This process is opposed by the strong electrostatic repulsion between the negatively charged phosphate groups. RNA molecules overcome this repulsion through the direct or indirect coordination of divalent metal ions. In group I self-splicing introns, a magnesium-organized ion core containing five magnesium ions constructs an exterior surface that facilitates the close packing of noncanonical loops into the minor groove of a double helix. Within the interior of this ion core, base-stacking and hydrogen-bonding interactions between nucleotides form specific metal ion binding sites (10, 11).
3. Quaternary Structure of RNA
There are relatively few well-characterized examples of the association of RNA molecules to form supramolecular quaternary structures, but these are relatively important. For example, during pre-mRNA splicing, messenger RNAs associate with five major ribonucleoprotein (RNP) particles called small nuclear RNPs; such snRNPs interact with each other and with mRNAs. These interactions and their dynamic disruption and formation by means of RNA-RNA quaternary interactions are essential for RNA splicing to occur (13).
In most examples characterized thus far, the quaternary association of RNA molecules occurs by conventional Watson-Crick base pairing. For example, small regulatory RNAs with longer complementary sequences within RNA molecules (antisense RNAs) form intermolecular duplexes during the control of gene expression in both prokaryotes and eukaryotes (14). Similarly, guide RNA recognizes complementary sequences to identify sites where mRNAs are edited post-transcriptionally (15) (see RNA Editing). Although more complex RNA quaternary structures do not rely exclusively on Watson-Crick pairing, base pairs are still very important. So-called "kissing-hairpins" form between self-complementary loop nucleotides in two stem-loop structures (16, 17). These structures provide protein-recognition sites during the regulation of prokaryotic plasmid copy number, and possibly during the dimerization of the HIV genome. The best-characterized example of supramolecular association of RNA molecules by means of non-Watson-Crick interactions is provided by so-called "G-quartet structures" (18). These structures form readily in vitro for RNA and DNA sequences containing stretches of guanidines or uracils (18), but it is not clear whether these structures occur at all in vivo.
4. Summary
In conclusion, formation of Watson-Crick base pairs provide a simple structural code of RNA secondary structure formation, and helical pairing can be predicted very successfully by phylogeny and thermodynamics. As demonstrated for the first time by tRNA, tertiary interactions between the secondary structural elements fold RNA molecules into their three-dimensional structures. Helical regions generally define the RNA secondary structure. Tertiary contacts between secondary structure regions of the RNA, often involving nucleotides in single-stranded regions, fold the RNA into its three-dimensional structure. RNA quaternary structures are less well understood, yet relatively important in gene expression and its regulation.