1. Domains and Motifs Found in RNA-Binding Proteins
The majority of the known RNA-binding proteins have modular structures that contain an RNA-binding domain combined with other auxiliary domains (1, 2). Four RNA-binding sequence motifs have been found in RNA-binding proteins from diverse species: therefore, it is considered that they arose early in evolution. These correspond to (1) the RNP domain, (2) the KH domain , (3) the dsRNA-binding domain, and (4) the S1 domain . X-ray crystallography has shown or NMR that they contain an a/b fold similar to those found in some ribosomal protein subunits, and it has been suggested that these RNA-binding motifs may have evolved from ribosomal proteins. This might help to explain why RNA-binding domains are generally distinct from DNA-binding domains. Notable exceptions are the zinc finger, OB fold, and homeodomain. It is an interesting observation that many known RNA-binding proteins are all b or a/b proteins that contain an exposed b-sheet. The crystalline structure of the complex between U1A protein and an RNA hairpin discussed later suggests why a b-sheet is a good RNA-binding surface.
1.1. RNP Domain
The RNP domain, also known as RNA recognition motif (RRM) or RNP-consensus sequence (RNP-CS) type RNA-binding domain, is found in more than 200 distinct RNA-binding proteins from diverse species (1, 2, 6). It is also the second most common protein sequence motif in the entire genome of the nematode Caenorhabditis elegans. These facts suggest that this module appeared early in evolution. The RNP domain is about 80 amino acid residues long and contains two highly conserved short sequence motifs called RNP octamer (RNP1) and RNP hexamer (RNP2). Some proteins contain only a single RNP domain, but others contain multiple copies. For example, the protein that binds to the polyadenylate (poly A) tail of mRNA in eukaryotes contains four tandem copies of the RNP domain.
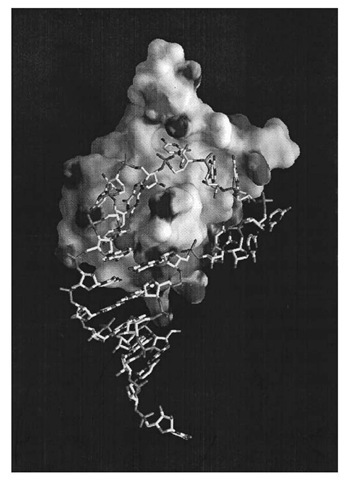
The three-dimensional structure of the RNP domain first determined for U1A spliceosomal protein, indicated that it consists of a four-stranded antiparallel b-sheet flanked on one side by two a -helices. The RNP1 and RNP2 motifs are located in the two middle b-strands of the sheet, and the side chains of the three highly conserved aromatic residues within RNP1 and RNP2 are projected onto the surface of the b-sheet (in U1A one of these residues is replaced by a glutamine) (7). U1A protein is a protein component of U1 snRNP, a large RNA/protein complex involved in pre-mRNA splicing, and binds to an RNA hairpin that contains 10 nucleotides in the loop. The crystal structure of a complex between U1A protein and an RNA hairpin representing its binding site has been determined at 1.9 A resolution (8). The RNA hairpin loop binds to the surface of the b-sheet as an open structure, and the first seven nucleotides of the loop are fitted into a groove formed on the surface of the b-sheet (Fig. 1). The polypeptide loop between the b2 and b3 strands protrudes through the RNA loop. The bases of the seven nucleotides are splayed, and the protein-RNA contacts are made almost exclusively by RNA bases. These seven bases stack onto an adjacent base, a protein side chain, or both and also form an intricate hydrogen-bond network with protein side chains and the amide and carbonyl groups of the protein main chain. U1A protein also binds to the 3′ -untranslated region of its own pre-mRNA and prevents its polyadenylation by directly interacting with poly-A polymerase. The binding site contains two internal loops, each with the AUUG(C/U)AC heptamer found in U1 snRNA hairpin II (6, 9, 10).
Figure 1. Crystal structure of a complex between U1A spliceosomal protein and its hairpin RNA binding site in U1 snRNA (8). The contact surface of the protein molecule is shown, whereas the RNA is a skeletal model. The AUUGCAC sequence in the ten-nucleotide loop fits into the groove on the surface of the b-sheet and binds tightly through stacking and hydrogen bond interactions with the protein.
1.2. Double-Stranded RNA-Binding Domain (dsRBD)
The dsRBD is a short sequence motif found in multiple copies in RNA-binding proteins from diverse origins including Escherichia coli, Drosophila, Xenopus, and mammals. In the Drosophila embryo, Staufen protein binds to maternal bicoid mRNA and plays an important role in establishing the anterior-posterior polarity through mRNA localization (11). E. coli ribonuclease III is an important enzyme involved in processing ribosomal and transfer RNA. The dsRBD is also found in adenosine deaminase, which is a key enzyme in RNA Editing, and in the double-stranded RNA-dependent protein kinase, which plays an important role in viral gene expression. The NMR structures of dsRBD from RNase III and Staufen protein show that the module contains a three-stranded antiparallel b-sheet that has two a-helices packed on one side (11, 12). The dsRDB shows strong similarities to the N-terminal domain of Bacillus stearothermophilus ribosomal protein S5 at the levels of both amino acid sequence and three-dimensional structure (11). This strongly suggests that dsRBD may have evolved from a ribosomal protein. It is not yet known how this domain binds RNA, but site-directed mutagenesis experiments suggest that loops between b1 and b2 and between b3 and a2 may be involved in RNA-binding. The major groove of dsRNA is narrow and deep, and the bases are not accessible by protein side chains for sequence-specific interactions. It is likely that each dsRBD binds to the minor groove or to the phosphate backbone in the major groove. Biochemical experiments suggest that a set of dsRDB, together with some auxiliary domains, may recognize a structural feature of folded RNA.
1.3. KH Domain
Heterogeneous nuclear ribonucleoprotein (hnRNP) K is one of the nuclear proteins that bind to precursors of mRNA (1). There are three copies of a sequence motif within hnRNP K, and homologous sequences have been found in many proteins from diverse species, from bacteria to man, that interact with RNA. Human Fragile X (FMR1) gene contains two copies of the KH domain. A mutation in one of these is responsible for severe hereditary mental retardation (1, 13). The natural RNA target of the FMR1 gene product has not been identified, but the mutation that causes severe hereditary mental retardation abolishes the binding of poly(U) to the FMR1 protein. This suggests that the ability of FMR1 to bind to RNA is essential for its in vivo function. Two proteins that contain three copies of the KH domain are associated with the 3′-untranslated region of a-globin mRNA. These proteins affect the in vivo half-life of the a-globin mRNA. Ribosomal protein S3 from E. coli and other bacteria contains the KH domain, suggesting that the KH domains found in higher organisms have evolved from a ribosomal protein (1). The NMR structure of the KH domain from human vigilin has an a/b structure that contains a three-stranded b-sheet with three a-helices (14).
1.4. S1 Domain
Ribosomal protein S1 from Escherichia coli contains six copies of a short sequence motif of approximately 80 amino acid residues. S1 protein is directly involved in RNA binding because it can be cross-linked to mRNA in the translational initiation complex. The S1 domain has been found in proteins from bacteria and eukaryotes, including polynucleotide phosphorylase, initiation factor 1 (IF1), NusA, ribonucleases II and E from E. coli, yeast RNA helicase PRP22, eukaryotic initiation factor eIF2a, and eIF2a kinase inhibitor. An NMR study of the S1 domain of polynucleotide phosphorylase shows that this domain consists of a five-stranded antiparallel b-barrel (15).
1.5. MS2 Bacteriophage Coat Protein
One of the best studied examples of an RNA-binding protein is the MS2 bacteriophage coat protein. The MS2 phage contains a genomic RNA 3569 nucleotides long, packaged into an icosahedral protein shell that consists of 180 copies of the coat protein. A capsid protein dimer binds to an RNA hairpin formed near the ribosomal-binding site of the virally encoded replicase gene, thereby inhibiting translation of the replicase mRNA. The interaction also triggers assembly of the coat protein and packaging of the genomic RNA. A synthetic RNA hairpin that represents the binding site was soaked into a crystal of the empty phage particle, and its structure was determined to 2.8 A resolution (16). The RNA, which contains a tetraloop and a bulged adenosine, binds to the surface of the continuous b-sheet across the dyad axis. The side chain of a tyrosine residue stacks onto a cytidine in the tetraloop, and the bulged adenosine and an adenosine in the tetraloop bind to equivalent sets of residues from each subunit.
1.6. Zinc-Containing RNA-Binding Proteins
Transcription factor IIIA is a transcriptional activator of the 5 S rRNA gene, but it also binds to its gene product 5 S to rRNA, and functions as a storage or transport protein (17). It contains nine zinc fingers, each of which folds into a domain that contains two b-strands and one a-helix stabilized by the coordination of two histidine and two cysteine residues to a zinc ion. Crystallographic analyses of zinc finger proteins in complex with dsDNA show that the a-helix fits into the major groove of the DNA and forms many sequence-specific contacts with the DNA bases (18). It is believed that 5 S rRNA folds into a branched double helix that contains many bulges which may widen the major groove of RNA to permit entry of the recognition helices. The HIV nucleocapsid protein is another example of a zinc-containing, RNA-binding protein. Others are E. coli alanyl-tRNA synthetase and tRNA guanine transglycosylase (28).
1.7. Other RNA-Binding Modules
The bases of the anticodon loops of tRNA Asp and tRNALys are splayed over the surface of a b-barrel domain of their cognate tRNA synthetases and are recognized in a sequence specific manner (see later) (19, 20). Many structural homologues of this b-barrel domain, which is known as the OB-fold, have been found in proteins that bind oligonucleotides (both ssRNA and ssDNA) or oligosaccharides (21).
The Sm proteins that form the core of spliceosomal small nuclear ribonucleoprotein particles (snRNP) contain a conserved sequence motif. It is predicted that this domain has an a/b fold, but its structure is yet to be determined (2). Some hnRNP proteins contain multiple repeats of an Arg-Gly-Gly (RGG) sequence that are believed involved in RNA binding (1).
The crystal structure of the SRP 14 / 9 heterodimer, which binds to the Alu domain of the mammalian signal recognition particle RNA, has been determined. SRP9 and SRP14 are structurally homologous and contain the same a-b-b-b-a fold, related to but distinct from the dsRNA-binding module. The heterodimer has pseudo two-fold symmetry and is saddle-like, comprising a strongly curved six-stranded amphipathic b-sheet. The four helices are packed on the convex side, and the exposed concave surface is lined with positively charged residues.
Both HIV tat and rev proteins bind the TAR and RRE elements, respectively, to their target RNA sequences, by arginine-rich peptides. The solution structure of a 14-residue arginine-rich peptide from BIV tat complexed with BIV TAR has been determined by NMR (26). The peptide forms a b-hairpin that interacts in the RNA major groove.
2. tRNA-Binding Proteins
2.1. Aminoacyl-tRNA Synthetases
The fidelity of protein synthesis depends to a large extent on the extreme specificity with which aminoacyl-tRNA synthetases charge their cognate tRNA with their cognate amino acid. In E. coli, there are at least 46 different tRNA molecules that have anticodons which correspond to the various amino acids. The seryl-tRNA synthetase, for example, has to charge the six serine isoacceptors (including one for selenocysteine) selectively and ignore the others. Because tRNA superficially have similar secondary and tertiary structures, what is the molecular basis for the specific recognition between aminoacyl-tRNA synthetases and tRNA? In many cases, extensive biochemical studies have revealed the so-called tRNA identity elements (see Aminoacyl tRNA Synthetases).
A more detailed picture of specific tRNA recognition and catalysis by aminoacyl-tRNA synthetases is emerging from crystallographic studies of aminoacyl tRNA synthetases complexed with various combinations of their three substrates: ATP, cognate amino acid, and cognate tRNA, plus the activated amino acid intermediate, the aminoacyl-adenylate. Now, crystal structures are known of 14 of the 20 aminoacyl tRNA synthetases, five of which are in complexes with cognate tRNA (3, 4). The three systems, for which the most extensive structural data exists on protein-tRNA recognition, are the class I glutaminyl system and the class II aspartyl and seryl systems. These show strikingly different modes of specific synthetase-tRNA interaction but share certain general features, including a fairly large synthetase-tRNA interactive interface characterized by (1) nonspecific backbone contacts, often involving basic residues, both of which increase binding affinity and aid correct positioning and orienting the tRNA; (2) discriminatory base-specific interactions restricted to a few regions, principally the anticodon and the acceptor stem. The second general feature is mutual induced fit by which protein-RNA contacts are made as a result of conformational changes in either or both macromolecules. This includes ordering of protein loops and reorienting and stabilizing domains (eg, SerRS), base-pair breaking in the acceptor stem, and 3′ -end distortion (eg, tRNAGln) and destacking of bases in the anticodon loop (eg, tRNAGln, tRNA Asp).
2.2. Glutaminyl-tRNA Synthetase (GlnRS)
GlnRS is a monomeric class I synthetase whose specificity for tRNAGln is largely determined by interactions with identity elements in the tRNA acceptor stem and anticodon stem-loop, both of which have severe distortions from the structure found in uncomplexed tRNA. In the E. coli complex, the tRNA anticodon stem is extended from five to seven base pairs by two extra non-Watson-Crick base pairs (23). The three anticodon bases (CUG) are splayed to fit into three separate recognition pockets formed at the interface between the distal two b-barrel domains of the protein. In the active site of the synthetase, the tRNA is oriented so that specific interactions can be made within the acceptor stem’s minor groove to identity determinants in base pairs 2 and 3. On the other hand, the tRNA 3′ -end reaches the catalytic center only by forming an unusual hairpin turn. This conformation requires breaking the first U1-A72 base pair and is stabilized, in part, by a hydrogen bond between the discriminator base G73 and the phosphate of A72.
2.3. Aspartyl-tRNA Synthetase (AspRS)
AspRS is a dimeric class IIb synthetase that binds two tRNAs symmetrically, although each tRNA interacts predominantly with only one subunit. In contrast to class I, class II synthetases interact with the acceptor stem of their cognate tRNA from the major groove side (27) by the so-called motif 2 loop, which in yeast AspRS makes base-specific interactions with the discriminator base G73 and base pair U1-A72 (19). The major groove side recognition also means that the single-stranded 3′ -end of the tRNA enters the synthetase active site without significant distortion from its normal helical path, again in strong contrast to class I. Anticodon recognition by AspRS is performed by an N-terminal, five-stranded, b-barrel domain (OB fold) (21). The normal compact structure of the free tRNA anticodon loop undergoes a large conformational change, and the five anticodon loop bases are exposed to the exterior. The three anticodon bases (GUC) lie across the b-sheet surface and are recognized by specific hydrogen bonding interactions (19). Recent crystallographic results on another closely related class IIb synthetase complexed with its cognate tRNA, T. thermophilus lysyl- tRNA synthetase, show a very similar interaction between the anticodon of tRNALys (CUU) and the N-terminal, b-barrel domain of LysRS (20). Whereas the central U35 interacts identically in the two systems (by stacking with a conserved phenylalanine residue and hydrogen bonding with conserved glutamine and arginine residues), the specificity for base 36 and to a lesser extent for base 34 is idiosyncratic for each synthetase.
2.4. Seryl-tRNA Synthetase (SerRS)
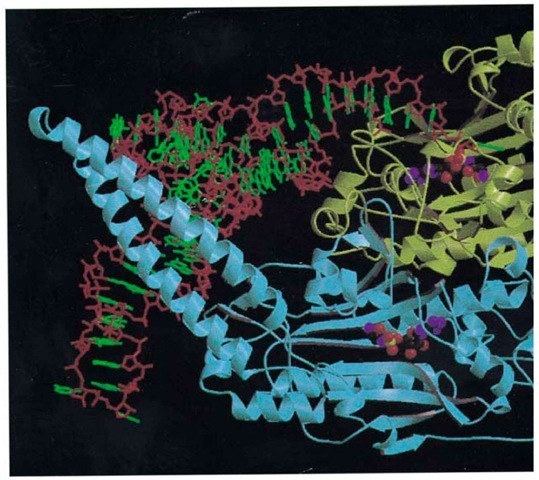
SerRS is also a dimeric class II synthetase, but its mode of tRNA recognition differs significantly from that of AspRS (Fig. 2). SerRS is characterized by a unique 100-residue, N-terminal domain that is folded into a 60 A long, solvent exposed and flexible, antiparallel coiled coil, known as the helical arm (24, 25). The variety of serine anticodons from two distinct groups means that the serine isoacceptors share no common anticodon base. Thus the anticodon is not an identity element of tRNASer, whereas the long variable arm is, a feature shared only by tRNA Leu and tRNATyr. The main features of the recognition of tRNA by T. thermophilus SerRS are (1) the tRNA binds across the two subunits of the dimer; (2) upon tRNA binding the helical arm of the synthetase is stabilized in a new orientation and binds between the TYC loop and the long, variable-arm of the tRNA; (3) contacts with the tRNA long, variable-arm backbone extend until the sixth base pair, explaining the need for a minimum length of the tRNA variable arm; (4) the synthetase makes many backbone contacts, but few base-specific interactions, and is principally recognizing the unique shape of cpr Qpr tRNA . The unique shape of tRNA is largely determined by bases 20A and 20B inserted into the D-loop, both of which play novel roles in tertiary interactions in the tRNA core. In particular, the base of G20B is stacked against the first base pair of the long variable arm and thus defines the spatial orientation of the latter; (5) the anticodon stem loop is not in contact with the synthetase; (6) the motif 2 loop of SerRS (longer than that of AspRS) makes base contacts down to the fourth base pair within the acceptor stem’s major groove, but they are, however, only weakly discriminatory (25).
Figure 2. Overall view of the ternary complex of seryl-tRNA synthetase, tRNASer(GGA), and a nonhydrolyzable analog monomer 1 is in yellow, and monomer 2 is in blue. Only the backbone and its secondary structure are shown (arrows are tRNA backbone is in red, and the bases are in green. The tRNA is viewed looking down the anticodon stem, which is not in the figure. The long variable arm of the tRNA crosses the helical arm of monomer 2 perpendicularly and emerges at th the Ser-AMS molecule is represented by spheres.
2.5. Elongation Factor EF-Tu
EF-Tu is a G-protein that, in the activated GTP-bound state, binds all aminoacylated elongator tRNA with higher affinity than uncharged tRNA and delivers them to the ribosome. The crystalline structure of the ternary complex of yeast tRNA Phe, T. aquaticus EF-Tu and GDPNP has recently been determined and reveals how activated EF-Tu contacts relatively limited and conserved regions of aa-tRNA (5). The only parts of the aa-tRNA in contact with EF-Tu are the aminoacylated CCA end, the 5′ -phosphate, and the T-stem helix. The long stem of the L-shaped tRNA projects away from the protein, so that the entire complex is extremely elongated. The CCA-3 end binds in a cleft between domains 1 and 2 of EF-Tu, and conserved residues from domain 2 interact with the base and phosphate of A76. Main-chain hydrogen bonds from EF-Tu are made to the ester group linking the carboxyl group of the amino acid to the 3′ -OH of the ribose and to the free amino group of the amino acid. The amino acid itself projects into a pocket formed between domains 1 and 2. The tRNA 5 ‘ -phosphate is bound by conserved basic residues from helix B of domain 1 and from domain 2. One side of the T-stem helix is packed against a depression between domains 1 and 3. The contacts are made by nonconserved residues from domain 3.
It has been pointed out that the EF-Tu.GTP.tRNA Phe ternary complex extraordinarily similar in shape to that of the elongation factor EF-G.GDP binary complex. This mimicry suggests that the two complexes might bind to the same ribosomal state.