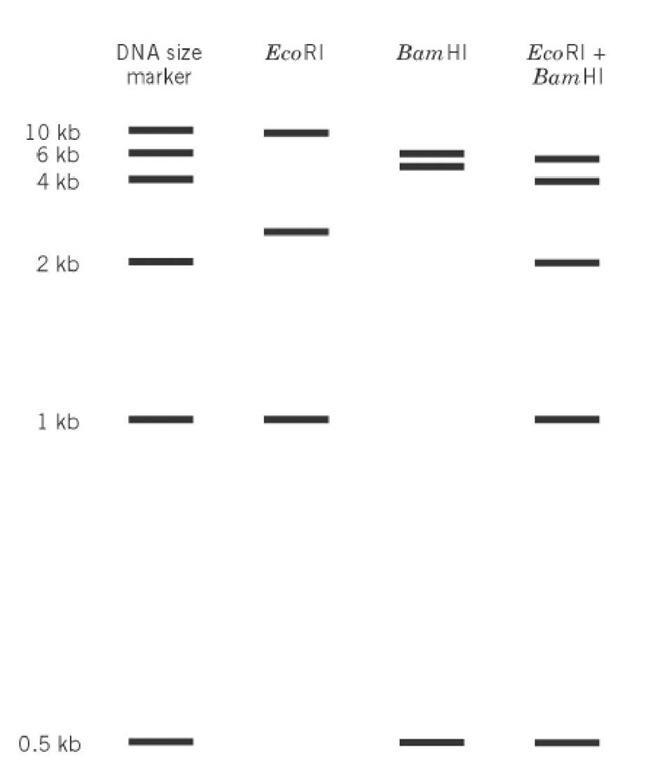
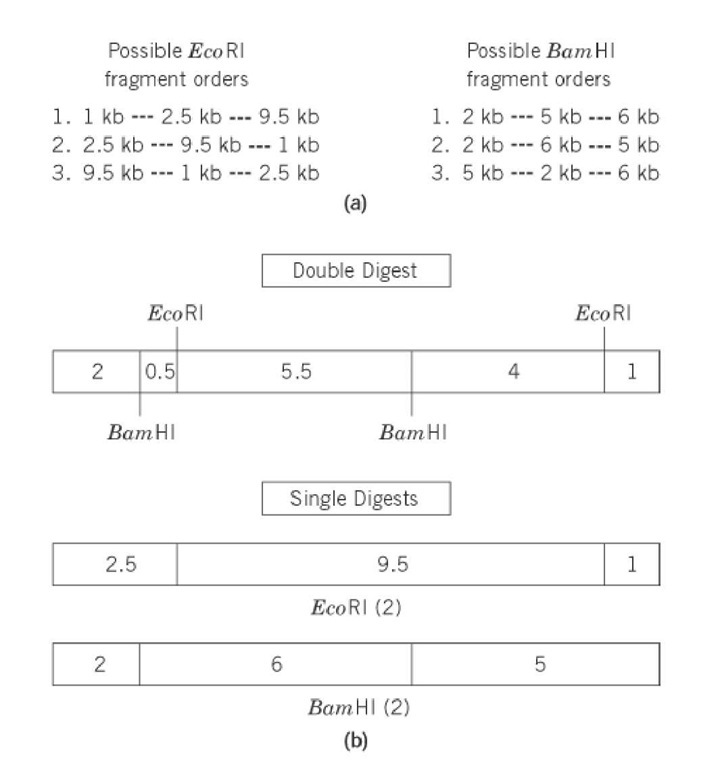
Restriction mapping is a technique for determining the linear order of restriction sites (see Restriction Enzymes) along a DNA molecule. The restriction fragments that are generated by digestion of DNA with different restriction enzymes, alone or in combination, and separated by gel electrophoresis, are used to deduce a restriction map. The resulting map gives the ordered arrangement of the restriction fragments. The choice of restriction enzyme, whether a rare or frequent cutter, depends upon the size of the DNA to be mapped and the frequency of its sites. For instance, rare-cutting restriction enzymes were used for initial mapping of large genomes like those of Escherichia coli and Homo sapiens, containing 4.2*106 base pairs and 3.3*109 , base pairs respectively. The resulting large restriction fragments isolated were further mapped by digestion with more frequently cutting enzymes. The fragments are separated by electrophoresis according to the inverse of their molecular weights, specifically, their lengths. Fragment size is determined by comparing the sample with a collection of standards. To illustrate, a linear 13-kb piece of DNA is mapped using two enzymes, EcoRI and BamHI. A diagram of the electrophoretic separation of the digests in an agarose gel is given in Figure 1. The DNA was digested with EcoRI, BamHI, and both enzymes. The sizes of fragments produced by single-enzyme digests indicate the number of restriction sites for each enzyme and the distances between them, while the fragments from the double digest indicate the relative distances between the BamHI and EcoRI sites (Fig. 2). EcoRI digestion produced three restriction fragments, 1, 2.5, and 9.5 kb in size. BamHI digestion also produced three fragments, 2, 5, and 6 in size. These findings indicate that each enzyme has two restriction sites on the linear DNA. A linear DNA will give ( n+1 ) fragments, where n is the number of restriction sites. The double digest reveals five restriction fragments—0.5, 1, 2, 4, and 5.5 kb in size—indicating four cleavages. The correct, consecutive ordering of fragments must be determined by reconciling each of the single-enzyme digests with the fragments arising from the double digest. All possible fragment orders for both enzymes are listed in Figure 2, as well as the order that gives the restriction map that agrees with the double-digest fragment sizes. In the example given, all fragment sizes from the two single digests and from the double digest sum to 13 kb each, the size of the original DNA. When determining a restriction map, one must remember that some fragments may be larger than the resolution power of the gel or that digestion may generate different fragments of the same size, which will migrate together. In such cases, alternative enzymes must be employed. Another approach uses the timed partial digestion of uniquely end-labeled, linearized DNA, which generates fragments ranging from the full-length DNA to the shortest fragment containing the label. The label identifies one end of the molecule. A restriction map can then be determined by a comparative analysis of the collection of fragments produced.
Figure 1. Idealized pattern of DNA fragments after digestion of a linear 13-kb DNA with EcoRI and BamHI, separately or combined, separation by electrophoresis, followed by visualization.
Figure 2. Restriction map derived from the data in Figure 1: (a) all possible combinations of linear restriction fragment orders from the individual EcoRI or BamHI digests; (b) diagram of the final restriction maps showing the correct, unique order of restriction fragments from a double digest with both EcoRI and BamHI of the 13 kb linearized DNA. Fragmentation patterns (2) of EcoRI and (2) of BamHI combine to give five fragments produced by the double-digest.
Restriction digestion breaks DNA into fragments of a manageable size for analysis (see Restriction Fragment). Restriction maps are thus helpful in determining gene order or chromosomal organization. Generation of restriction maps from different organisms reveals similarities or differences in their genomic arrangements. One important application of restriction mapping is genetic fingerprinting: the typing of individual DNA molecules based on differences in restriction map patterns. The differences in map patterns are reflected in restriction fragment length polymorphisms (RFLPs), which result from changes in restriction sites or distances between restriction sites due to local sequence variations. In some cases, RFLPs can be used as genetic markers for diagnosis of diseases, and can aid in isolating the gene or identifying DNA sequence aberrations causing the disease. Applications for analysis and manipulation of restriction fragments are discussed under the topic restriction fragment.