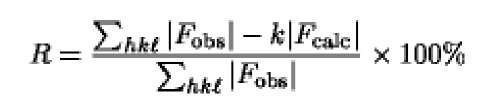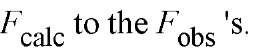The final result in X-ray crystallography is presenting a molecular model of the structure being determined. If this were an ideal model and if the packing of the models in the crystal structure were accurately known, the calculated the amplitude of the structure factor ^calc would be equal to the observed amplitude Fobs for each reflection. In practice this ideal situation does not exist, and the difference between the two values indicates model’s inadequacies. This difference, averaged over all of the measured reflections, is expressed as the crystallographic R-factor or reliability or residual index R:
or alternatively
where k is a factor scaling the
If the atoms were to be placed entirely at random in a non-centrosymmetrical structure like a protein, R would have the value 0.59. For correct structures refined to, for example, 2.0 A resolution, the R-factor is of the order of 0.20. If it is much higher, the structure is not likely to be correct.
A better estimate for the reliability of a protein structure is the free R-factor (1). It is calculated for a random selection of 5 to 10% of the observed reflections. These reflections are not used in the refinement, so refining and testing are completely independent.