The quaternary structure describes the way two or more polypeptide chains are arranged within the same protein molecule. The term, suggested by J. D. Bernal, was used first by J. Kendrew in 1959 [(1)] to further develop the already classical hierarchy of K. Linderstrom-Lang. If (1) primary structure refers to the amino acid sequence, (2) secondary structure is alpha-helices or beta-sheets, and (3) tertiary structure is the fold of the polypeptide chain, quaternary structure refers to the assembly of several polypeptide chains, as in four-chain hemoglobin, or of many chains in virus capsids or muscle fibers. In 1959, Kendrew had just solved the crystal structure of the single-chain myoglobin, which illustrated secondary structure as the a-helix and produced the first image of a tertiary structure. Soon afterward, M. Perutz obtained the crystal structure of hemoglobin, making quaternary structure directly visible in atomic detail. We now have many more examples in proteins ranging from enzymes to transcription factors and membrane components.
Proteins with more than one polypeptide chain are oligomeric or multimeric, and their chains are also called subunits. Oligomeric proteins may be built of identical subunits, when they are called homo-oligomers and their quaternary structure can be coded in the formula an to indicate that they are made of n copies of an a subunit. Other oligomeric proteins are referred to as hetero-oligomers. The quaternary structure formula anbm describes a hetero-oligomer that contains two types of subunits a and b in m and n copies, respectively. Gtpases involved in cellular signal transduction are abg hetero-trimers. Mammalian hemoglobins may be written as either a 2b2 or (ab) 2. Immunoglobulins G also have this formula, but unlike the a and b chains of hemoglobin, the heavy and light chains of an immunoglobulin are covalently linked by disulfide bonds. a2 dimers and a4 or a2b2 tetramers are the most common formulae in globular oligomeric proteins. More complex quaternary structures do exist, however, especially in membrane proteins. Thus, the formula a2bgd describes the quaternary structure of the nicotinic acetylcholine receptor of the neuromuscular junction, which comprises four different chains with unequal stoichiometries.
In order to write a formula such as a2b2, we need to know the number and type of the constituent polypeptide chains. This cannot be deduced from just the knowledge of the gene sequences, although genetic evidence may point to the presence of more than one type of chain. Determination of the formula usually requires expressing and purifying the protein. After purification, SDS-PAGE or mass spectrometry are commonly used to separate the individual polypeptide chains and estimate their apparent molecular weights. If a homogeneous protein yields more multiple species with different molecular weights, it is probably a hetero-oligomer, but quantitative peptide sequencing will be needed to determine the relative amounts of each chain type and to eliminate the possibility that they derive from proteolysis or post-translational modification of a single gene product.
Whether or not the constituent polypeptide chains are of one or several types, their actual number in the protein is ascertained by measuring the molecular weight of the native protein complex. The most commonly used methods are gel filtration and sedimentation equilibrium ultracentrifugation. However, neither is as easy and sensitive as SDS-PAGE, and both are much less accurate than mass spectrometry, two techniques that yield the molecular weights of the components, but not of the assembly. Moreover, the results from gel filtration and ultracentrifugation may be misinterpreted if the assembly is not fully stable, so that partial dissociation or aggregation takes place under conditions where the measurement is made. Chemical crosslinking followed by SDS-PAGE is an alternative to molecular weight determination. Covalent crosslinking an a n protein can, in principle, yield n bands on the gel having apparent molecular weights corresponding to species![]()
However, not all the constituent chains may form crosslinks readily, and the conditions where a complete ladder of n bands can actually be observed may be difficult to obtain. When the quaternary structure does not dissociate spontaneously during a separation, the number of subunits can be determined by hybridizing two distinguishable forms of the subunit, say, A and B, and then separating the various oligomers on the basis of this difference. For a tetramer, this should yield five species: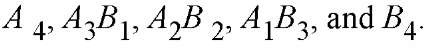 In practice, molecular weight determination is more difficult and less reliable for a native protein than for a denatured protein. Indeed, we lack that basic information for the vast majority of the proteins known only as putative gene products or bands on a gel. In addition, quaternary structures are sometimes revised, and tabulated values should be verified.
In practice, molecular weight determination is more difficult and less reliable for a native protein than for a denatured protein. Indeed, we lack that basic information for the vast majority of the proteins known only as putative gene products or bands on a gel. In addition, quaternary structures are sometimes revised, and tabulated values should be verified.
Beyond counting polypeptide chains, the quaternary structure is part of the three-dimensional structure as a whole. NMR and X-ray crystallography studies can determine the symmetry relationships between subunits, the location of their interfaces, and the nature of their atomic contacts, together with the fold of the polypeptide chains. In crystals, contacts between subunits coexist with intermolecular, crystal packing contacts. The subunit contacts can usually be recognized just on the basis of their greater extent, but ambiguities sometimes arise. They must be resolved by other methods, such as the site-directed mutagenesis of the proposed subunit contacts. To characterize the contacts by NMR, special isotope labeling techniques have been developed. In a dimer, one subunit is labeled with C, the other with N, and isotope-edited NOE signals are specifically detected between CH hydrogen atoms of the first subunit and NH hydrogen atoms of the other. However, the large sizes of oligomeric proteins generally make the NMR resonance lines broad, so the spectra are poorly resolved and difficult to assign. Indeed, neither NMR nor crystallography can dispense with the determination of the molecular weight of native protein.
The subunit contacts that make up the quaternary structure are generally noncovalent and form interesting examples of specific protein-protein interactions. They almost always make a major contribution to the stability of the native protein and often its function also. There are many oligomeric proteins in which a ligand-binding site exists at the interface between subunits. This is the case of the antigen-binding sites of immunoglobulins and of the DNA-binding sites in the great majority of DNA-binding proteins. Enzyme active sites at subunit interfaces are not uncommon, and allosteric mechanisms of regulation largely rely on properties of the quaternary structure. These aspects are discussed under Oligomeric proteins, together with the evolutionary aspects of quaternary structure.
