Inorganic pyrophosphatase (PPase) was first discovered in animal tissues in 1928 (1). It hydrolyzes pyrophosphate (PPj) to two molecules of inorganic phosphate (Pj). Soluble cytoplasmic PPase (s- PPase) is believed to be both ubiquitous and essential (2-4), playing a central role in cell metabolism by hydrolyzing the truly staggering quantity of PPj produced as a by-product of the biosyntheses of nucleic acids, proteins, lipids, and polysaccharides. Klemme (5) calculated that bacterial PPj levels would rise to 3 M in an hour in the absence of PPase, and several kilograms of PPj are generated in adult humans per day. The required high flux of PPj hydrolysis results from the high specific activity of s-PPase [turnover numbers at 25°C and neutral pH of ~100500 s-1 (6, 7), 1010 faster than the uncatalyzed rate (8)] and its relative abundance, typically 0.1-0.5% of cell protein by weight.
PPase catalysis of PPj hydrolysis drives biopolymer synthesis (9), but the high rate of PPj production results in steady-state cellular PPj concentrations that in some cells are well above equilibrium levels. Thus, at conditions close to physiological, the concentration of PPj in equilibrium with 10 mM Pi is about 0.1 mM (10, 11), whereas in bacterial, plant, and yeast cells PPj levels of the order of millimolar or greater are observed (5, 12, 13). Even in mammals, where the PPj concentrations are generally much lower, millimolar levels have been found in blood platelets (14), starved hepatocytes (15), and hepatocytes metabolizing acetate in the presence of Ca2 (16).
The steady-state levels of PPj can be important for metabolic regulation. For example, there is a linear relationship between the PPj concentration and the error rate in DNA polymerization (17) (see DNA Replication), and a model for PPj regulation of the termination of transcription in Escherichia coli has been presented (18). A disorder of PPj metabolism is suspected as a cause of calcium pyrophosphate dihydrate crystal deposition disease, a major arthropathy in the elderly that has also been linked to osteoarthritis (19, 20). Although as yet no linkage has been established between these diseases and a PPase abnormality, s-PPase has been shown to be effective in dissolving calcium pyrophosphate dihydrate crystals (21).
In addition to s-PPase, plants and certain bacteria have a membrane-bound PPase, structurally unrelated to s-PPase, which works as a reversible proton pump. In these organisms, the PPi level is much higher, and PPi is used as a phosphoryl and energy donor instead of, or in parallel with, ATP(22, 23). Mitochondrial PPase is a third kind of PPase, which, though structurally and functionally similar to s-PPase (3), contains noncatalytic subunits that anchor it to the inner mitochondrial membrane.
s-PPases have been studied from a large number of organisms (6, 22). Here we focus on the two best understood s-PPases, those of Saccharomyces cerevisiae (Y-PPase) and E. coli (E-PPase). Both of these enzymes have been cloned and expressed, and the structural and functional properties of wild-type enzyme, as well as many variants, have been characterized (7). Comprehensive recent reviews of E- and Y-PPases may be found in Refs. 6, 24, and 25.
1. Substrate Specificity
Isolated PPase requires added divalent metal ion for activity. Only four of many divalent ions tested have been shown to confer PPase with appreciable (>5% of maximal) PPj hydrolysis activity, with relative activities in the order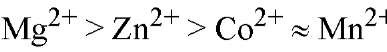 " (26). E-PPase shows similar specificity toward divalent metal ions (27). For Y-PPase, the lower effectiveness of
" (26). E-PPase shows similar specificity toward divalent metal ions (27). For Y-PPase, the lower effectiveness of![]() and
and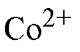 as cofactors for
as cofactors for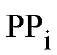 hydrolysis than
hydrolysis than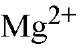 is due mainly to the slower rates of product
is due mainly to the slower rates of product![]() release from enzyme in the presence of each of these ions.
release from enzyme in the presence of each of these ions.
With![]() + as cofactor, the reaction is almost totally specific for
+ as cofactor, the reaction is almost totally specific for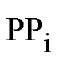 as substrate; in contrast, with
as substrate; in contrast, with 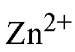 and/or
and/or![]() , other pyrophosphates, such as ATP and serial pyrophosphate, are hydrolyzed at rates within a factor of 10 of that of
, other pyrophosphates, such as ATP and serial pyrophosphate, are hydrolyzed at rates within a factor of 10 of that of![]() itself (28-30).
itself (28-30).![]() is not a substrate, but is a potent competitive inhibitor of PPase, with a K considerably lower than the Km ( Michaelis constant) of
is not a substrate, but is a potent competitive inhibitor of PPase, with a K considerably lower than the Km ( Michaelis constant) of 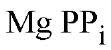 under corresponding conditions (31). In the presence of
under corresponding conditions (31). In the presence of![]() , high specificity is also evident for the back reaction from
, high specificity is also evident for the back reaction from![]() to synthesize enzyme-bound
to synthesize enzyme-bound![]() Thus, enzyme catalysis of thiophosphate, phosphoramidate, or fluorophosphate hydrolysis, which would be analogous to
Thus, enzyme catalysis of thiophosphate, phosphoramidate, or fluorophosphate hydrolysis, which would be analogous to![]()
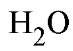 oxygen exchange (Eq. (1)), proceeds very slowly if at all. Fluoride shows very potent inhibition of Y-PPase (but not E-PPase), by formation of a stable, isolatable complex: PPase-
oxygen exchange (Eq. (1)), proceeds very slowly if at all. Fluoride shows very potent inhibition of Y-PPase (but not E-PPase), by formation of a stable, isolatable complex: PPase- (32).
(32).
Paralleling its substrate specificity, PPase displays a high binding specificity for its natural ligands, PPi and Pj. The two Pj sites per PPase subunit differ in affinity. Both the higher-affinity (P1) and the lower-affinity (P2) sites bind Pj more tightly as a function of added metal ion (33). Some changes are tolerated for the atom bridging the phosphoryl groups. In the presence of Mg , both O3PCHOHPO3 and O3PNHPO3 are reasonably good competitive inhibitors, although O3PCH2PO3 is not. With Mn2+, the specificity is somewhat relaxed: PPPj has a Km similar to that of PPj, and O3PCHOHPO3 binds quite tightly. The binding of analogues of Pi has been examined only in the presence of Mn .
None of the analogues tested, methyl phosphonate, phosphoramidate, or thiophosphate, bind comparably to Pj in site P1, but methyl phosphonate binds comparably to Pj in site P2. Thus, site P1 appears the primary source of binding specificity (33).
The outstanding features of Equation 1 are as follows: (a) All three forward rate constants, k3, k5, and k7, are partially rate-determining for PPj hydrolysis; (b) synthesis of enzyme-bound PPj from product Pi proceeds quite readily, but release of bound PPi (step 2) is very slow; (c) the equilibrium
2. Kinetic Mechanism
Y-PPase and E-PPase share a common mechanism for catalysis of PPj hydrolysis, PpH^O oxygen exchange, and PP^Pi exchange (34, 35). PPj hydrolysis proceeds via single-step direct phosphoryl transfer to water, without formation of a phosphorylated enzyme intermediate (Eq. (1)):
constant for hydrolysis of enzyme-bound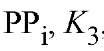 , is only 4-5, in contrast to much higher value for the hydrolysis of PPj in solution; and (d) the first Pj released (step 5) is the electrophilic
, is only 4-5, in contrast to much higher value for the hydrolysis of PPj in solution; and (d) the first Pj released (step 5) is the electrophilic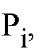 containing an oxygen atom from the nucleophilic water (34). The value of
containing an oxygen atom from the nucleophilic water (34). The value of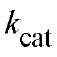 goes through a maximum as a function of pH (at pH 8 for E-PPase, 7 for Y PPase), implying that both acidic and basic groups are important for activity (7, 36). The value of
goes through a maximum as a function of pH (at pH 8 for E-PPase, 7 for Y PPase), implying that both acidic and basic groups are important for activity (7, 36). The value of also displays a pH optimum near neutrality for each enzyme.
also displays a pH optimum near neutrality for each enzyme.
3. Structural Studies on PPases
Y-PPase is a homodimer (286 amino acid residues per subunit), typical of eukaryotic PPases (37), and E-PPase is typical of prokaryotic PPases in being a homohexamer (175 residues each) (38). Although the enzymes have only about 27% sequence identity (6), they have a very similar protein structure, consisting of eight beta-strands and two alpha-helices (39) and belonging to the nucleotide-binding motif. The E-PPase subunit may be thought of as embedded in the Y-PPase subunit, and the common parts of the two molecules are best described as a distorted, highly twisted five-stranded b-barrel with four excursions, the latter creating a very large active site. About 14 polar active-site residues, highly conserved in s-PPases, are significant for catalysis (7), and 12 of them are in the excursions. In addition to E-PPase and Y-PPase, more than a dozen soluble PPases have been cloned and sequenced (6, 24).
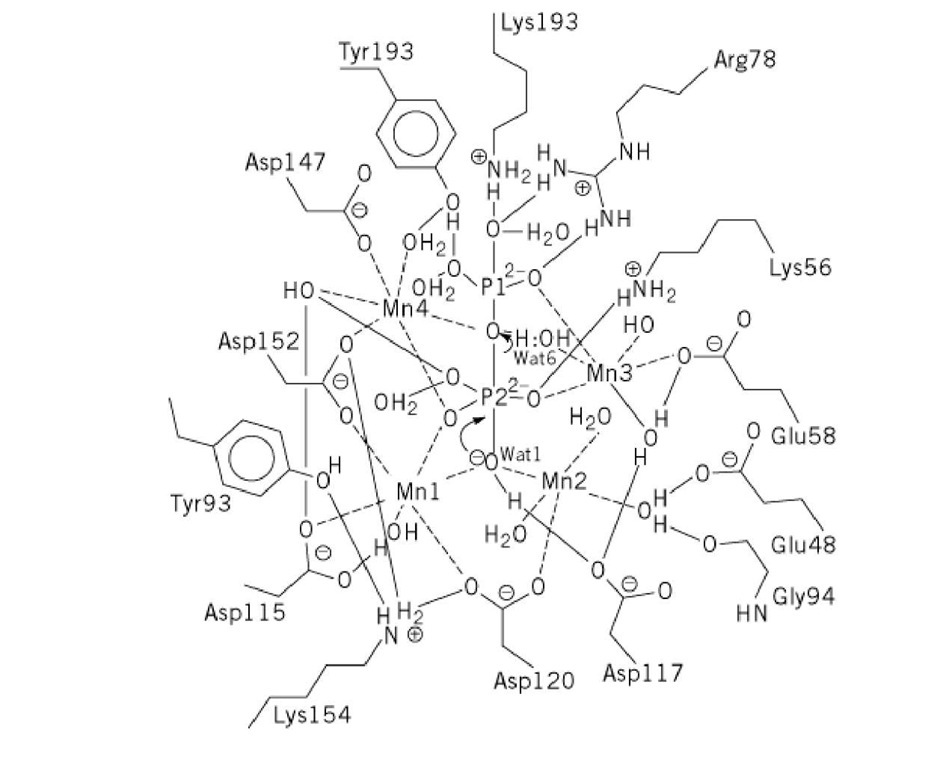
High-resolution structures have been determined for the Mgj 5 (39) and Mg2 5 (40) complexes of E-PPase and for the Mn2 and Mn4(P^2 complexes of Y-PPase (25, 41), where the numbers refer to the stoichiometries of ligand per monomer. These ligands are all bound at the active site, with the exception of the half Mg site in E-PPase. The latter reflects Mg binding at the trimer:trimer interface, which stabilizes the enzyme but otherwise has little effect on enzyme activity (42). In these structures, the E-PPase active site residues are virtually superposable on those of Y-PPase, demonstrating the highly conserved nature of the PPase active site. The 2.2-A resolution structure of the product Mn4(P^2 complex of Y-PPase (25) has been the most useful for considerations of mechanism (see text below).
4. Chemical Mechanism
PPase is presently unique among phosphoryl transfer enzymes in having four divalent metal ions at the active site, although it is probable that not more than three of these are required for hydrolysis (43). The structure of the M^^^ complex of Y-PPase, along with functional studies of wild-type and variant PPases, allows formulation of a detailed proposal for the catalytic mechanism (Fig. 1). In the proposed mechanism, PPase catalyzes PPj hydrolysis by lowering the pKa of the leaving group in site P1 (studies of phosphoryl transfer reactions in solution show that the rates of such reactions increase by 10- to 20-fold for each unit lowering of the leaving group pKa), by forming an incipient hydroxide ion that functions as a stronger nucleophile than water, and by shielding the charge on the electrophilic phosphorus in site P2, thus permitting attack by the hydroxide anion. According to this mechanism, the leaving phosphoryl group is activated by coordination to Arg78, Tyr192, Lys193, Met3, Met4, and Water6, the pKa of the nucleophilic water is lowered by coordination to Met1, Met2, and Asp117, and the charge on the electrophilic phosphoryl group is neutralized through coordination to Lys56, Tyr93, and Met1 to Met4. The extraordinary array of Lewis acids (four divalent metal ions; Lys, Arg, and Tyr side chains, bound waters) within the active site activate PPj hydrolysis through coordination to almost all of the available electron pairs in the oxygens of bound PPi. The essential base implicated by both steady-state and pre-steady-state pH-dependent studies is assigned to the presumptive nucleophilic water, Wat1, which has an abnormally low pKa (~ 6) by virtue of its coordination to Met1, Met2, and Asp117. These same studies implicate the essential acid in a step following PPi hydrolysis, which is believed to be protonation of the phosphate in site P1 prior to its dissociation from enzyme (44).
Figure 1. Mechanism and active-site structure of Y-PPase as deduced from the crystal structure of the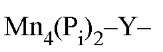 PPase complex.
PPase complex.
Interestingly, the proposed PPase mechanism has much more in common with those proposed for inorganic phosphatase and DNA polymerases and exonucleases than with that proposed for ATPase, which catalyzes a chemically more analogous reaction (25). Studies of the crystal structures and functional properties of critical active-site variants, such as Arg78Lys and Asp117Glu, that test various aspects of the proposal in Figure 1, are underway.
![tmp7-34_thumb[1] tmp7-34_thumb[1]](http://what-when-how.com/wp-content/uploads/2011/05/tmp734_thumb1_thumb.jpg)