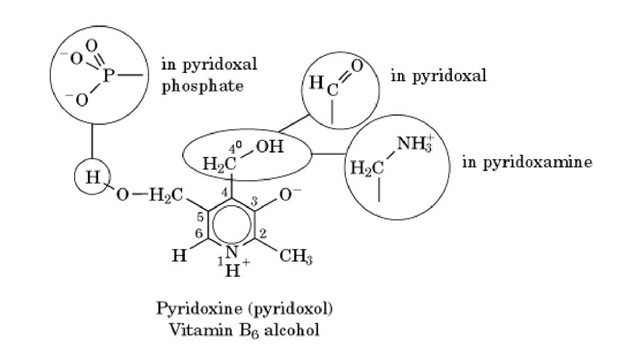
Pyridoxal 5′-phosphate (PLP), a phosphate ester of the aldehyde form of vitamin B6, is an essential coenzyme for numerous Enzymes (Table 1), many of which function in all living cells. PLP is a rather simple derivative of 3-hydroxypyridine. Its structure and that of related forms of vitamin B6 are as follows: (Structure 1) Within the human body, dietary vitamin B6 is converted into the coenzyme forms PLP and pyridoxamine 5′-phosphate (PMP). For all these compounds, the dipolar ionic ring structure shown predominates in solution and is also found in most enzymes.
Table 1. Groups of Enzymes Utilizing Pyridoxal or Pyridoxamine Phosphate as a Coenzyme®
Removal of alpha hydrogenas H+ Aminotransferases
Aspartate aminotransferase (14, 15) D-Amino acid aminotransferase (16) Branched chain aminotransferase (17) Gamma-aminobutyrate aminotransferase (18) Serine:pyruvate aminotransferase (19) Alanine racemase (20)
Aminocyclopropane carboxylate synthase (21) 2-Amino-3-oxobutyrate-CoA ligase (22) AKB Synthase Beta elimination and replacement reactions L-and D-Serine dehydratases (deaminases) (23, 24) Tyrosine phenol-lyase (25) Alliinase (26)
Cystathionine b-lyase (cystathionase) (27) O-Acetylserine sulfhydrylase (cysteine synthase) (28) Tryptophan synthase (29-32) Removal of alpha carboxylate as CO2 (33)
Diaminopimelate decarboxylase (34)
Glycine decarboxylase (requires lipoyl group) (35)
Glutamate decarboxylase (36, 37)
DOPA decarboxylase (38)
Dialkylglycine decarboxylase (39)
Removal of side chain by aldol cleavage
Serine hydroxymethyltransferase (40)
Reactions of ketimine intermediates Threonine synthase (3) Other enzymes
Lysine 2,3-aminomutase (41) Glycogen phosphorylase (42)
Pyridoxamine phosphate (PMP) in synthesis of 3,6-dideoxy-L-arabino-hexose (43)
a Names of a few representative enzymes are also listed.
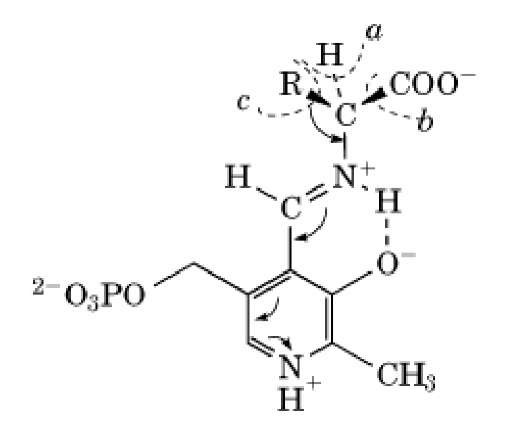
Within the active sites of most PLP-dependent enzymes, the carbonyl group of the coenzyme forms a Schiff Base with an amino group of a specific lysine side chain. This Schiff base is commonly called the internal aldimine. When an amino acid substrate binds into the active site, a series of rapid reactions occur, many of which have been characterized spectroscopically and kinetically (1-6). The amino group of the substrate is deprotonated and then adds to the carbon atom of the C=N Schiff base group of the internal aldimine, forming an intermediate geminal diamine . After another proton transfer, elimination of the lysyl side-chain amino group leads to a stable hydrogen-bonded Schiff base with the substrate, the external aldimine: (Structure 2) In this structure, the protonated ring nitrogen provides a powerful electron-accepting center that can assist in breaking any one of the three bonds marked a, b, and c on Structure (2), as indicated by the curved arrows (7). Which type of reaction occurs is determined by the structure of the enzyme active site and the way in which the amino acid part of the Schiff base is held. The bond to be cleaved must be nearly perpendicular to the plane of the coenzyme ring and to the conjugated p-electron system of the Schiff base. (8) Side-chain cleavage (c) occurs only for b-hydroxyamino acids and related compounds.
Sell iff base
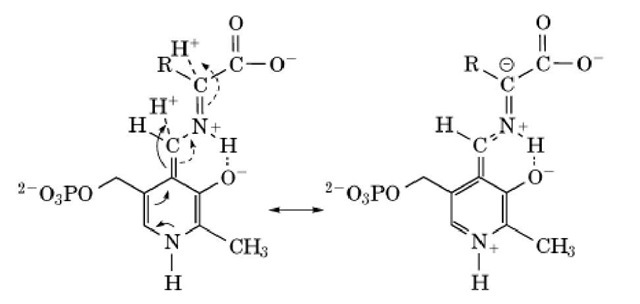
The product of the bond cleavage has been called the quinonoid or carbanionic intermediate, and the product of removal of H+ by a catalytic base is illustrated with cleavage of bond a: (Structure 3) The two resonance structures shown illustrate the quinonoid and carbanionic attributes. The carbanionic structure is stabilized by the adjacent positively charged hydrogen-bonded proton. Another resonance form has the negative charge on the 4′ carbon rather than the a carbon. It is possible that the proton on the Schiff base nitrogen may be shifted onto the phenolate oxygen of the ring in this intermediate (9). There are still uncertainties about the details of the reaction mechanism.
Quino m>i d- carban ionic structure
The quinonoid-carbanionic form shown in Structure (3) can undergo reprotonation at either Ca or C4′. Protonation at Ca, but without stereospecificity, leads to racemization of the amino acid (see Stereoisomers) and is observed for bacterial alanine and glutamic acid racemases. Transaminases (aminotransferases) catalyze protonation at either Ca (with retention of the configuration) or C4′. The initial product in the latter case is another Schiff base called the ketimine: (Structure 4) Its hydrolysis produces pyridoxamine phosphate (PMP), the product of a half-reaction of a transaminase.
Ketimine
Removal of the a-hydrogen is also a first step in a number of other reactions, such as synthesis in plants of the ethylene precursor aminocyclohexane carboxylate and numerous reactions involving elimination of the b or g substituent. For example, the bacterial tryptophan indole-lyase releases indole from tryptophan, the remainder of the molecule being converted to pyruvate and ammonia. The g-elimination reactions are more complex and pass through a ketimine step before the elimination.
Removal of the a-carboxylate as carbon dioxide is characteristic of a large number of decarboxylases, including glutamate and dihydroxyphenylalanine (DOPA) decarboxylases of the brain. Cleavage of the side chains is of more limited occurrence but accomplishes another series of important metabolic reactions. Best known of these is cleavage of serine by serine hydroxymethylase to give glycine and formaldehyde. The latter does not leave the active site but is captured by tetrahydrofolate as methylenetetrahydrofolate.
There are many medical and technological aspects to the study of PLP. Amino acid racemases are targets for antibacterial drugs, and several enzymes are targets for herbicides and pesticides. Enzyme deficiency diseases are known. Poor removal of homocysteine leads to homocysteinemia, which is linked to atherosclerosis (10, 11). PLP has some blocking effect on the virus HIV (12). PLP is widely used as a labeling reagent for amino groups in proteins by reduction of Schiff bases formed with borohydride (13), which may be radiolabeled with deuterium or tritium.