Photoreactivation is the reversal of the effects of ultraviolet (UV) light (200 to 300 nm) by simultaneous or subsequent exposure of the organism to near UV-visible (300 to 500 nm) light (1, 2). Photoreactivation prevents the induction by UV light of growth delay, mutation, and cell death. Although several factors contribute to the phenomenon of photoreactivation, the main mechanism involved is the reversal of UV-induced pyrimidine dimers in DNA by the enzyme called photoreactivating enzyme, or DNA photolyase (3). DNA photolyase is the only known enzyme to employ light energy to catalyze a chemical reaction (3).
The two major lesions induced in DNA by UV light are cyclobutane pyrimidine dimers (Pyr<>Pyr) and [6-4] photoproducts (Pyr[6_4]Pyr) (Fig. 1). There are separate photolyases that are specific for Pyr<>Pyr and for [6-4] photoproducts (4, 5), but no known enzyme can act on both lesions. The structures and reaction mechanisms of the two types of photolyases are similar and may be summarized as follows. Each consists of a single polypeptide chain of 55 to 65 kDa containing a chromophore that acts as a photoantenna (folate or deazaflavin) and a catalytic cofactor (FADH 2) (6-9). It binds to the photodamage in DNA in a light-independent manner, producing a relatively stable enzyme-substrate complex. Upon exposure to light, the enzyme breaks the bonds that link the two pyrimidines to one another, generates standard canonical bases as products, and then dissociates from the repaired DNA. In contrast to many other enzymatic reactions that involve electron transfer, catalysis by photolyase involves a cyclic electron transfer and is not a redox reaction, in that upon completion of the enzymatic reaction the redox states of the enzyme and the substrate remain the same (6).
Figure 1. The two major photoproducts produced in DNA by ultraviolet light from (a) thymine dinucleotide: ( b) the cyclobutane thymine dimer, and (c) the [6-4] photoproduct. Similar photoproducts arise from other adjacent dipyrimidines in DNA.
1. Evolution
Photolyases are flavoproteins, yet the primary sequences of the apoenzymes do not reveal any obvious homology to the signature sequences found in other flavoproteins (4). Instead, photolyases have remarkable sequence homology to blue-light photoreceptors (10), which regulate circadian rhythms in organisms ranging from mustard to humans (11-13). It appears that the necessity for the flavin cofactor to function from the light-excited state has imposed unique constraints on the flavin binding site of photolyase/blue-light photoreceptor family of proteins, such that the active-site geometry of these proteins is different from all other flavoproteins that carry out catalysis with ground-state flavin.
The distribution of the members of the photolyase/blue-light photoreceptor family in the biological world does not fit a readily defined pattern of evolutionary selection. The coliform bacterium Escherichia coli contains photolyase, but the soil bacterium Bacillus subtilis does not. The budding yeast Saccharomyces cerevisiae has the enzyme, but the fission yeast S. pombe does not. Drosophila melanogaster possesses cyclobutane pyrimidine dimer photolyase, [6-4] photolyase, and blue-light photoreceptor (14), while humans lack photolyase but express two blue-light photoreceptor proteins. Table 1shows the expression pattern of this class of proteins in select members of the "three kingdoms of life": bacteria, archaea, and eukarya. No general rules emerge from this table. The only rules that appear to have arisen from investigations of these groups of proteins are the following. (i) Cyclobutane photolyase has been detected in bacteria, plants, and animals through marsupial animals (15), but it has not been found in placental mammals such as human (16) and mouse. (ii) The [6-4] photolyase has been found in plants, insects (Drosophila), and some vertebrates (Xenopus, rattlesnake), but not in mammals such as human or mouse, nor in bacteria. (iii) Blue-light photoreceptors have been found in plants (Arabidopsis, mustard) and animals (Drosophila, human, and mouse), but not in bacteria. (iv) Of all bacterial and eukaryotic viruses investigated, photolyase has, so far, been found only in a grasshopper virus.
Table 1. Distribution of the Photolyase/Blue-Light Photoreceptor Family of Proteins in the Biological World
|
Bacteria |
Archaea |
Eucarya |
||||
|
E. coli |
B. subtilis |
M. thermoauto |
M. janaschii |
S. cerevisiae |
D. melanogaster |
H. sapiens |
|
Photolyase Yes |
No |
Yes |
No |
Yes |
Yes |
No |
|
[6-4] No |
No |
No |
No |
No |
Yes |
No |
|
Photolyase |
||||||
|
Cryptochrome No |
No |
No |
No |
No |
Yes |
Yes |
The tissue distribution of photolyase in animals that possess the gene is rather surprising. It is highly expressed in Drosophila ovaries, where it is expected to protect germ cells from UV damage. However, it is also expressed in all internal organs of opossum, including high levels of expression in the brain. Considering the nocturnal nature of the opossum, and the low probability even in bright daylight of UV and photoreactivating light penetrating the skull of the opossum, this expression pattern remains enigmatic.
2. Structure
Photolyases consist of a single polypeptide of 500 to 700 amino acid residues and two noncovalently bound cofactors/chromophores. One of the chromophores is always flavin adenine dinucleotide (FAD), and the other is either a pterin (methenyltetrahydrofolate in the case of E. coli and S. cerevisiae photolyases) or deazariboflavin. Deazariboflavin is considered an ancient molecule, and photolyases with this cofactor have been found only in a few organisms that are capable of synthesizing this uncommon cofactor: Aspergillus nidulans, Streptomyces griseus, and M. thermoautotrophicum. Most other photolyases characterized to date, including those from E. coli, S. cerevisiae, D. melanogaster, and marsupials, contain a pterin as the second chromophore. Only two [6-4] photolyases have been characterized in detail, those of D. melanogaster and X. laevis, and both contain a pterin as the second chromophore (17). Blue-light photoreceptors from mustard, A. thaliana, and humans contain FAD and a pterin (11, 12).
The structures of E. coli and A. nidulans photolyases have been determined (18, 19). Even though the two enzymes are only 39% identical in sequence, they have remarkably similar structures, such that the traces of C a; atoms are superimposable. The enzymes consist of two well-defined domains: an N-terminal a/b domain, which is connected to the C-terminal alpha-helical domain through a long inter-domain loop. The folate or the deazariboflavin chromophores are located in a crevice separating the two domains. In the E. coli enzyme, the folate is close to the surface of the protein and hence is not tightly bound to the enzyme; furthermore, its orientation is not optimal for energy transfer to the FAD cofactor. In contrast, the deazariboflavin chromophore in A. nidulans photolyase is deeply buried between the two domains, and it has a more favorable orientation relative to FAD for efficient energy transfer between the chromophores. Thus even though center-to-center distances between the two chromophores in the two photolyases are about 17 A, the energy transfer from the second chromophore to FAD is more efficient (97% to 70% ) in A. nidulans photolyase than in that of E. coli (20). The helical domain is made up exclusively of a-helices that are packed in two rather compact clusters. The FAD is buried between these two clusters, and the flavin and adenine rings of the cofactor are in the unusual cis conformation in both photolyases.
The crystal structures of both enzymes also reveal how they bind to DNA. A plot of the surface potential of the enzyme reveals a longitudinal groove traversing both the a/b and the a-helical domains that is paved with positive charges. At its center is a hole that has the shape and size appropriate for cyclobutane pyrimidine dimer. The bottom of the hole is made up by the flavin ring. These structural features suggest that the DNA lies in the positively charged groove through ionic interactions between the DNA backbone and the positive charge of the side chains and that the dimer "flips out" of the duplex into the hole within the enzyme. Following repair, the flipped-out dinucleotide would undergo a conformational change upon transformation of the dimer into two monomers, which would force the bases out of the hole and assist the repaired DNA to dissociate from the enzyme. Biochemical data indicate that [6-4] photolyase also employs a "flip-out" mode of complex formation during catalysis.
3. Reaction Mechanism
Photolyase works by the classical Michaelis-Menten kinetic mechanism; that is, the enzyme and substrate form a noncovalent complex that is converted to an enzyme-product complex after catalysis and eventually dissociates to yield product and free enzyme. The light-dependence of this catalysis step is unique to photolyase.
3.1. Binding
Photolyase, unlike some other high-specificity DNA-binding proteins, does not rely on diffusion along the linear DNA molecule to recognize its substrate. Similarly, the enzyme, in contrast to most proteins that bind to DNA in a sequence-specific manner and hence require double-stranded DNA, binds to its cognate lesions in single- and double-stranded DNA with almost equal affinities (21). This property of the enzyme, combined with the fact that the enzyme can bind the photoproduct even in a dinucleotide form and repair it with high affinity, are consistent with a model in which the enzyme binds mostly to the backbone of the damaged strand and flips-out the photolesion into the catalytic hole for repair.
3.2. Catalysis
A most extraordinary aspect of catalysis by photolyase is its absolute dependence on light. According to the Woodward-Hoffman rule on symmetry in molecular orbitals, the symmetry of molecular orbitals in a chemical reaction is conserved between reactants and products. A corollary of this rule is that photochemically allowed reactions that proceed from the lowest unoccupied molecular orbitals (LUMO) are thermally forbidden; and conversely, thermally allowed reactions that proceed from the highest-occupied molecular orbitals (HOMO) are photochemically forbidden. In line with these rules, the cyclobutane ring of Pyr<>Pyr is stable at pH 1 and 160 °C for 6 h, but it is reversed to pyrimidine monomers with 240-nm light with a quantum yield of about 1.0. Surprisingly, however, even though photolyase utilizes light energy to catalyze the repair, the reaction is not a photochemical reaction in the strict sense, because it does not involve photoexcitation of the pyrimidine dimer, either directly or indirectly. Consequently, splitting of Pyr<>Pyr by photolyase does not break the rule of conversation of orbital symmetry. The enzyme accomplishes this feat by converting the Pyr<>Pyr into a radical, which then readily proceeds, thermally, to monomers. Light (300 to 500 nm) is used simply to excite the cofactor of the enzyme into a strong reductant capable of donating an electron to the relatively inert Pyr<>Pyr.
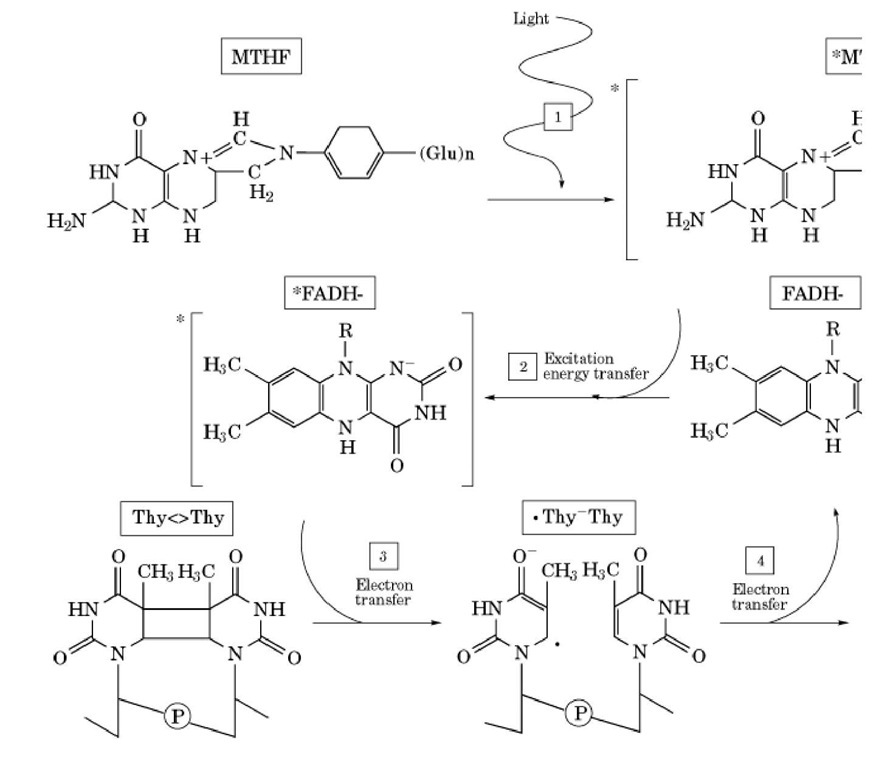
Having thus stated the general features of catalysis, the specifics for Pyr<>Pyr photolyase can be summarized as shown in Figure 2. The enzyme binds to its substrate independent of light, and the reaction proceeds upon exposure to light as follows (6). First, a photon is absorbed by the photoantenna (pterin or deazaflavin) to excite the chromophore to the excited singlet state. Second, the photoantenna transfers the energy by dipole-dipole interactions to FADH2 with either 70% (pterin) or 97% (deazaflavin) efficiency. Third, the excited singlet state of FADH2 transfers an electron to Pyr<>Pyr with about 100% efficiency, generating flavin neutral radical and Pyr<>Pyr radical anion. Fourth, the Pyr<>Pyr radical anion splits to pyrimidine monomers, concomitant with electron transfer back to restore the flavin to its catalytically active state. The reaction is a photoinitiated cyclic electron transfer, which results in bond rearrangement without changing the redox status of the reactants.
Figure 2. Photoreactivation photocycle for photolyase. The active site of the enzyme complexed to substrate is shown. ( pterin) absorbs a near-UV photon. (2) The excited singlet-state pterin transfers energy to FADH- by dipole-dipole intera FADH transfers an electron to the thymine dimer (4). The thymine dimer anion radical collapses to two thymines, conc< the flavin neutral radical.
The [6-4] photolyase functions in a similar manner (17, 22), with one important exception. Formation of [6-4] photoproduct encompasses joining of C6 of the 5′ base to the C4 of the 3′ base via a sigma bond, accompanied by transfer of the group at the C4 position (-NH or -OH) of the 3′ base to the C5 position of the 5′ base. Thus, simple cleavage of these new bonds would not restore the bases to normal. Hence, before the photoinduced cleavage of these bonds, the enzyme converts the "open form" of [6-4] photoproduct to an oxetane intermediate in which the NH or OH that was transferred to the C5 position of the 5′ base is shared by the C5 of the 5′ base and the C4 of the 3′ base (22). When the oxetane intermediate receives an electron from the active-site flavin, the resulting radical intermediate collapses to the two original bases, with back electron transfer to restore the flavin radical to its catalytically active form (Figure 3).
Figure 3. [6-4] Photolyase photocycle. Upon binding to the enzyme, the open form of the [6-4] photoproduct is convert photocycle proceeds as follows: (1) The photoantenna absorbs a photon and (2) transfers energy to the catalytic cofactor, 4] photoproduct; (4) the photoproduct radical splits into two normal bases.
The blue-light photoreceptors that exhibit 40% to 60% sequence identity to photolyases (10) have absolutely no repair activity (11). They are photoreceptors for regulating plant growth or for photoentrainment of the circadian clock in animals (13). However, the reaction mechanism of these molecules is not known at present.
![The two major photoproducts produced in DNA by ultraviolet light from (a) thymine dinucleotide: ( b) the cyclobutane thymine dimer, and (c) the [6-4] photoproduct. Similar photoproducts arise from other adjacent dipyrimidines in DNA. The two major photoproducts produced in DNA by ultraviolet light from (a) thymine dinucleotide: ( b) the cyclobutane thymine dimer, and (c) the [6-4] photoproduct. Similar photoproducts arise from other adjacent dipyrimidines in DNA.](http://what-when-how.com/wp-content/uploads/2011/05/tmp10918_thumb_thumb.jpg)
![[6-4] Photolyase photocycle. Upon binding to the enzyme, the open form of the [6-4] photoproduct is convert photocycle proceeds as follows: (1) The photoantenna absorbs a photon and (2) transfers energy to the catalytic cofactor, 4] photoproduct; (4) the photoproduct radical splits into two normal bases. [6-4] Photolyase photocycle. Upon binding to the enzyme, the open form of the [6-4] photoproduct is convert photocycle proceeds as follows: (1) The photoantenna absorbs a photon and (2) transfers energy to the catalytic cofactor, 4] photoproduct; (4) the photoproduct radical splits into two normal bases.](http://what-when-how.com/wp-content/uploads/2011/05/tmp10920_thumb1_thumb.jpg)
