Only a few cis/trans isomerizations are catalyzed by enzymes. Most of these enzymes catalyze isomerization of C—C double bonds. However, the highly abundant and ubiquitously distributed peptidyl prolyl cis/trans isomerases (PPIases) have evolved to accelerate the rotation about a formal single bond. They catalyze the cis/trans isomerization of imide peptidyl-proline bonds and are inactive toward the -CONH- peptide moiety. In enzyme nomenclature, PPIases are classified under EC number 5.2.1.8.
Initially, PPIases were identified in the cytosol of kidney cells from their ability to accelerate the cis to trans isomerization of chromogenic tetrapeptides of the type succinyl-Ala-Xaa-Pro-Phe-4-nitroanilides (where Xaa is any natural amino acid) by an assay that utilizes the conformational specificity of chymotrypsin (1). Briefly, in this cis/trans assay, the trans prolyl bond isomer of the substrate succinyl-Ala-Ala-Pro-Phe-4-nitroanilide (about 92% of the total) is cleaved completely at the anilide bond in a few seconds when high concentrations (»rnM) of the proteinase are used. The remaining cis isomer of the peptide (8%) is resistant to chymotrypsin until it undergoes cis to trans isomerization. When monitored by the absorbance of the released product (4-nitroaniline), the isomerization is the rate-limiting step for proteolyzing the cis isomers indirectly. It can be quantified as a first-order reaction with a half-time of about 90 s at 10°C. This kinetic phase, which is equivalent to a quasi-irreversible cis to trans isomerization, has an accelerated rate in the presence of catalytic amounts of a PPIase. The calculated rate constants depend linearly on the PPIase concentration. This assay could not demonstrate catalysis in both directions, but this was possible with NMR (2, 3). The standard assay was greatly improved by dissolving the assay peptides in LiCl/trifluoroethanol to increase the fraction of cis isomer (4).
PPIases were initially anticipated to be folding/rearrangement catalysts for proteins bearing cis prolyl bonds in the native state because the slow trans to cis interconversion following protein synthesis on the ribosome will seriously limit the rate of in vivo protein folding . Indeed, PPIases increase the rate of the slow kinetic phases in in vitro refolding of many denatured proteins known to be limited in rate by prolyl isomerization. In support of a physiological role in protein folding, the involvement of a PPIase in triple-helix formation in type I pro-collagen was shown in chick embryo fibroblasts in cellulo (5). However, the biological functions of PPIases are not yet well understood, and they may have more general roles than affecting only de novo protein folding.
According to our current knowledge, the enzyme class of PPIases comprises three families with unrelated amino acid sequences: cyclophilins (abbreviated Cyp), FK506-binding proteins (abbreviated FKBP ), and parvulins. An additional subfamily, the prokaryotic trigger factors, exhibits weak sequence similarity to FKBP proteins but lacks FK506-binding ability (6). The amino acid sequences of several dozen different PPIases are known, encompassing molecular masses from 10.1 kDa (92 amino acids in length) for E. coli parvulin (7) to 158 kDa for the NK-TR1 cyclophilin of human, large granular lymphocytes (8). Typically, Escherichia coli cells harbor two cyclophilins, four FKBP-like PPIases, the archetype of parvulins, along with the SurA protein containing two parvulin domains, and the ribosome-bound trigger factor. Among the 470 predicted coding regions in the free-living bacterium Mycoplasma genitalium, which has the smallest known genome of any free living organism, a trigger factor homologue is the sole PPIase present (9).
Members of the PPIase families resemble perfectly evolved enzymes for catalytic efficiency (10). For oligopeptide substrates, both the Michaelis constant, Km, and the turnover number, kcat, are large and yield values of ,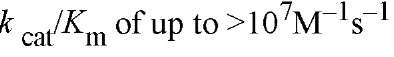 These bimolecular rate constants approach the diffusion controlled limits for enzyme reactions (see Enzymes). With
These bimolecular rate constants approach the diffusion controlled limits for enzyme reactions (see Enzymes). With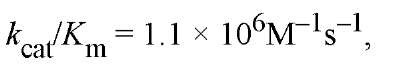 the E. coli trigger factor catalyzing refolding of a denatured ribonuclease T1 variant exemplifies a particularly efficient catalysis in protein folding (11). This efficacy results from having extended secondary binding sites to bind the unfolded protein substrate favorably. These subsites encompass the two protein domains flanking the central PPIase domain of the enzyme. Many of the PPIase enzymes are specific only for certain amino acid residues flanking the proline in the primary structure, but this is less important with the cyclophilins. Conversely, a hydrophobic side chain of the amino acid preceding the proline enhances catalysis by FKBPs.
the E. coli trigger factor catalyzing refolding of a denatured ribonuclease T1 variant exemplifies a particularly efficient catalysis in protein folding (11). This efficacy results from having extended secondary binding sites to bind the unfolded protein substrate favorably. These subsites encompass the two protein domains flanking the central PPIase domain of the enzyme. Many of the PPIase enzymes are specific only for certain amino acid residues flanking the proline in the primary structure, but this is less important with the cyclophilins. Conversely, a hydrophobic side chain of the amino acid preceding the proline enhances catalysis by FKBPs.
PPIases applied in catalytic amounts cannot alter the cis/trans equilibrium of the substrates, as expected. When protein refolding is hindered by misfolding and protein aggregation, an increase of the yield of native protein is not obtained by supplementing the refolding buffer with a PPIase. Thus, an important aspect of chaperones is missing in PPIases (12).
Immunosuppressive compounds, like cyclosporin A, FK506 and rapamycin, bind tightly to the respective active site of cyclophilins and FKBPs and inhibit their enzymatic activities reversibly and competitively and have inhibition constants in the picomolar to micromolar range. Cross-inhibition of the different classes does not occur. At the present time, no specific reversible inhibitors for parvulins and trigger factors have been reported.
Definitive identification of the natural substrates of PPI-ases has not been reported yet. Nevertheless, a number of binding proteins, which may include the putative cellular substrates, were identified by chemical cross-linking, affinity chromatography, the yeast two-hybrid screen, and by co-purification. For example, the nuclear parvulin-like PPIase Pin1 has affinity for NIMA protein kinase in the two-hybrid system. The catalytic activity of this isomerase is involved in cell-cycle control in eukaryotes (13). For the trigger factor family, association with nascent chains derived from secreted and cytosolic proteins and specific affinity for the 50S subunit of the E. coli ribosome are indicated by cross-linking and fractionation of enzyme activity (6, 14, 15). On the other hand, the same PPIase has been detected as an indispensable component of the heterooligomeric complex of the chaperone GroEL with the degradable fusion protein CRAG. This system has been used to determine the requirements for rapid degradation of abnormal, misfolded proteins by the proteinase ClpP in E. coli cells (16). When a trace of copurifying protein of the ryanodine receptor was analyzed, it became obvious that intracellular calcium release channels on the endoplasmic or sarcoplasmic reticulum (ryanodine receptor, RyR) and the inositol 1,4,5-triphosphate receptor constitute heterooligomeric complexes of the type ((RyR) 4/(FKBP12)4) with either cytosolic FKBP12 or FKBP12.6 (17, 18). A dysfunctional state, associated with the impairment of the receptor-mediated Ca retardation of the channel, arises from inhibiting PPIase activity with both FK506 and rapamycin (19). Furthermore, FKBPs and cyclophilins in the range of 40 to 60 kDa were found as components of the unactivated steroid receptor complex, associated with the hsp90 component of the receptor (20). Interestingly, two members of different families of proteins that assist protein folding, an enzyme and a chaperone, coexist as binding partners in this receptor complex.
