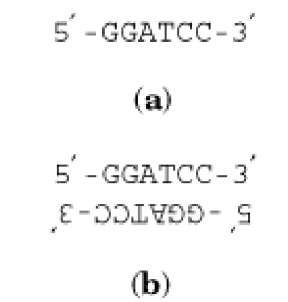A palindromic word or text reads the same in both directions. Strictly speaking, this definition applies only to the same line of the text. The term "palindrome" is traditionally applied to double-stranded DNA (which is "two lines") when the nucleotide sequence is read forward from one strand and backward from the other, complementary strand. The reading proceeds in the same direction in terms of chemical polarity, 5′ to 3′, because DNA is an antiparallel duplex. A more appropriate name for such double-stranded palindromes that is also frequently used is a "sequence or element with complementary symmetry."
The palindromes in the double-stranded sense as discussed above are not only built of two identical sequences, but they also have complementary symmetry (twofold rotational symmetry). One example, the recognition site for the Bam HI restriction enzyme, is shown in Fig. 1. Such a peculiar structure is typical of restriction sites. It guarantees that both strands—not just one—will be cut by the enzyme, either simultaneously or after a second approach by the enzyme. Nucleotide sequences of various lengths with complementary symmetry are rather common. For example, they comprise the consensus sequences of many repressor-binding sites, or operators (1).
Figure 1. A nucleotide sequence with complementary symmetry ( a) and a double-stranded palindrome (b)—the element with twofold rotational symmetry. Hydrogen bonds between the strands in (b) are not shown.
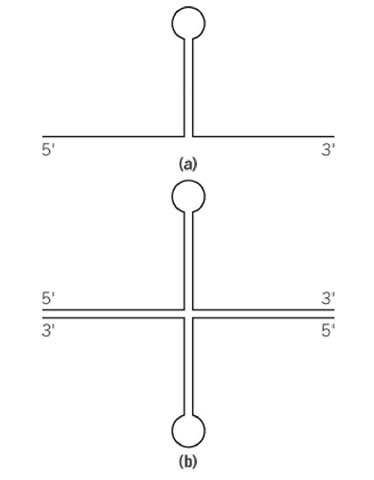
The condition of identity and complementarity of the two strands of the palindrome also makes the halves of each strand complementary to one another, with the potential for each strand to fold back on itself, forming a hairpin structure. In a palindromic DNA duplex, such hairpins could be made in both strands, so that a cruciform structure is formed (2) (see Fig. 2). The loops of the cruciforms generally could be any size and not necessarily complementary symmetric as the sequences in the stems of the cruciform. Another obvious property of palindromes is the identity of their double-stranded halves, except for their being placed in opposite polarity, like in inverted repeats, but without interruption between the repeats. (Various types of repeats are compared in the scheme presented in Tandem Repeats.) Generally speaking, any head-to-head (or tail-to-tail) fusion of identical fragments of a double-stranded DNA would generate a palindrome. For example, very long palindromes are formed in the process of gene amplification (3), in the size range of many kilobases. Complementary symmetries generally separated by rather long interruptions are characteristic of many transposable elements (4). A whole family of transposable elements also exists: the foldback element transposons, which are about 1 kb in size. These transposons are essentially made of inverted repeats with a very small middle loop region—almost a palindrome.
Figure 2. Schematic presentation of a hairpin structure in a single-stranded DNA or RNA (a) and of a cruciform structure (b). The double lines correspond to antiparallel complementary strands in the duplex parts of the structures.
Palindromes that read the same in both directions of the same strand, ie, proper (or true), palindromes also are known. These sequences possess a mirror symmetry, for example, ATGAGTA. Such elements are frequently encountered in intervening sequences, or introns (5), and may be interpreted as a device to avoid complementary folding of the sequence. Indeed, the absence of complementary sequence elements in the mirror-symmetric sequences of the true palindromes like ATHTA and TGHGT causes the sequences to be located in the single-stranded regions of the RNA secondary structure.
Another class of mirror-symmetry palindromes are the polypurine and polypyrimidine stretches in the H-form structure (6, 7). This partially triplex structure is formed in a normal DNA duplex at lowered pH or under torsional stress. The locally separated polypyrimidine strand folds back and binds to the duplex by Hoogsteen hydrogen bonds. The sequence of this third strand can be incorporated into the structure only if it is mirror-symmetric to the sequence of the polypyrimidine strand of the Watson-Crick duplex.