The oskar is a maternal effect gene in Drosophila melanogaster whose primary function is to organize a specialized cytoplasm, termed the germ plasm or pole plasm, at the posterior pole of the maturing egg (1). This germ plasm contains factors required for abdomen formation and germ-cell formation. The oskar messenger RNA is synthesized during oogenesis. It is deposited into the egg, where it becomes localized to the posterior pole through sequences in its 3′-untranslated region (UTR). Only posteriorly localized RNA is translated into Oskar protein. The site of Oskar protein synthesis determines where germ plasm is assembled, and the quantity of Oskar protein determines how many germ cells form. oskar is a member of the posterior group of genes (named for their effect on development of the abdomen in the posterior half of the embryo). Most of the posterior group genes facilitate localization and translation of oskar mRNA, while a small number of posterior genes act together with Oskar in the assembly of the germ plasm or are specifically required for either germ cell or abdomen formation.
1. Oskar Protein and RNA Structure
Oskar encodes a 2.9-kb mRNA that is expressed in the germ line of the adult female and is present at the posterior pole of early embryos. oskar mRNA has a 14-nucleotide 5′ UTR and a 1045-nucleotide 3′ UTR. Two different in-frame start codons are used to produce Oskar protein isoforms of 606 amino acids residues (69 kDa) and 468 residues (54 kDa). The short isoform has full oskar activity, although the long isoform is necessary to maintain localization of the oskar mRNA at the posterior of the oocyte (2, 3). The 3′ UTR contains regulatory elements for RNA localization and translation, while the region between the two start codons contains sequences necessary for translational regulation (4). The Oskar protein is novel, in that no known sequence or structural motifs are present. The only known homologue is from Drosophila virilis (5).
2. Oskar RNA Localization and Translation
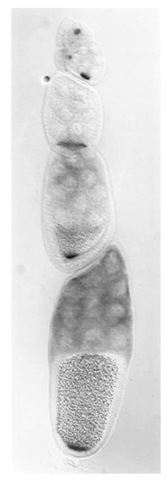
Oskar protein synthesis is regulated at the level of mRNA localization and translation and occurs during oogenesis. Each Drosophila ovary consists of approximately 16 ovarioles, each of which contains a series of developing egg chambers. At the tip of each ovariole, a germ-line stem cell produces a new stem cell and a cystoblast by asymmetric cell division. Each cystoblast undergoes four rounds of division with incomplete cytokinesis, giving rise to 15 nurse cells and the oocyte. The nurse cell-oocyte cluster is interconnected by cytoplasmic bridges called ring canals and surrounded by somatic follicle cells. oskar RNA is produced in the nurse cells and deposited into the developing oocyte, where it accumulates from the earliest stages of oogenesis. By late stage 8 of oogenesis, the RNA is concentrated at both the anterior margin and the posterior pole of the oocyte. By stage 10, oskar RNA is localized exclusively to the posterior pole, where it remains until it is degraded in the early embryo after germ-cell formation [see Fig. 1 (6, 7)]. Oskar protein is translated once the mRNA becomes localized to the posterior pole, while unlocalized RNA is translationally repressed. This ensures that protein is made only at the posterior pole of the oocyte and embryo (2, 3).
Figure 1. A Drosophila melanogaster ovariole. oskar mRNA is detected by in situ hybridization. In the earlier stages of oogenesis (top of figure), oskar mRNA accumulates in the oocyte. As each egg chamber matures, the oskar mRNA is localized within the oocyte to the posterior pole (center and bottom of figure). Anterior is to the top, posterior is to the bottom.
The site where oskar RNA is localized and Oskar protein is synthesized determines where germ cells form and the posterior body region, the abdomen, develops. Therefore processes that lead to the localization of oskar RNA are directly linked to the establishment of anterior-posterior polarity within the oocyte. All aspects of oskar RNA localization are mediated by sequences within the oskar 3′ UTR (8). Early aspects of oskar RNA transport into the oocyte and localization to the posterior pole of the oocyte require the correct organization of the microtubule network. Genes involved in the polarization of the early microtubule network that extends from the nurse cells to the oocyte also affect the transport of oskar into the oocyte. These genes include egalitarian, Bicaudal-D, (coiled-coil domain protein), orb (an RNA-recognition motif protein with similarity to mammalian CEBP), and cytoplasmic dynein (9). Subsequently, the establishment of anterior-posterior polarity within the oocyte is controlled by cell-cell signaling between the oocyte-bound Gurken protein, a member of the transforming growth factor-a (TGF-A) family, and its receptor, the Drosophila EGF receptor (DER or torpedo). Mutations in either gene disrupt localization of oskar RNA to the posterior pole, presumably by altering the polarity of the microtubule network within the oocyte. Indeed, inhibitors of the microtubule, but not of the microfilament, cytoskeleton disrupt the localization of oskar RNA during oogenesis (10). Other maternal effect genes that affect oskar localization and affect the microtubule organization in the oocyte include cappuccino (for min homologue), spire, and mago nashi (a novel protein that is highly conserved from plants to humans). A role of microfilaments in oskar mRNA localization is suggested by the effect that mutations in the tropomyosin II gene (an actin-binding protein) have on oskar RNA localization (11). It has been proposed that the actin cytoskeleton is required for high levels of oskar mRNA localization or the firm anchoring of the RNA at the posterior pole (11).
Staufen, a double-stranded RNA-binding protein, has been proposed as the best candidate to mediate directly the localization of oskar RNA. staufen mutants do not affect the organization of the cytoskeleton, and Staufen protein and oskar RNA colocalize during oogenesis (12). A direct physical interaction between Staufen and oskar RNA has not been demonstrated. Staufen is also required for localization of bicoid mRNA to the anterior of the oocyte and ofprospero RNA to the apical side of dividing neuroblasts (12).
Oskar mRNA that is not localized to the posterior pole is translationally repressed. If the RNA is translated prematurely, posterior body patterning will be specified throughout the embryo. Translational repression is mediated through specific RNA-binding sites in the 3′ UTR of oskar mRNA. The RNA-binding protein Bruno (which corresponds to the arrest gene independently identified on the basis of its early arrest maternal effect phenotype) binds specifically to Bruno response elements (BREs) in the oskar RNA (13). The bicaudal-C protein (a KH-domain protein) has also been implicated in repressing translation of unlocalized oskar RNA. Only at the posterior pole, where oskar RNA is localized, is the RNA translationally active. Activation of translation depends on derepression mediated through sequences between the two alternative start codons of the oskar mRNA (4). Translation of oskar RNA or the stability of Oskar protein is further dependent upon the gene aubergine as well as upon the Vasa protein, an RNA helicase that interacts physically with Bruno protein (14, 15).
3. Role of oskar in Germ Plasm Assembly
Homozygous mutant oskar females are viable, but produce embryos that lack germ plasm. These embryos fail to form germ cells and do not develop an abdomen. Mislocalization of the oskar coding region (via the RNA localization element in the 3′ UTR of bicoid mRNA) to the anterior of the oocyte results in the assembly of ectopic germ plasm, a mirror image duplication of the abdomen, and formation of ectopic germ cells (16). Thus oskar is both necessary and sufficient for assembly of germ plasm and for formation of both the abdomen and germ cells.
Assembly of the germ plasm is also dependent upon other factors that act downstream of oskar. Vasa is a DEAD-box helicase that physically interacts with Oskar. The products of the tudor and valois genes are also required for germ plasm assembly. Vasa, Tudor, and Oskar are components of electron-dense particles, known as polar granules, that are found only in the germ plasm (17).
The effect of oskar on abdomen formation is mediated through the nanos gene. Oskar is responsible for localizing nanos mRNA to the posterior pole of the embryo, where Nanos protein is translated and acts to facilitate abdomen formation through repression of the hunchback mRNA. While the mechanism of nanos localization to the posterior pole is not clear, oskar is both necessary and sufficient for nanos localization. Overexpression of oskar or mislocalization of the oskar mRNA results in an excess of nanos activity at the site of oskar localization.
Oskar also recruits several factors to the posterior pole that play a role in germ-cell formation. These include: germ cell-less mRNA, which encodes a protein that associates with the nuclear envelope; mitochondrial large ribosomal RNA (mtlrRNA) that is found outside of the mitochondria, where it associates with polar granules during the early stages of embryogenesis; and polar granule component 1 (pgc1), which codes for an untranslated RNA. A number of different experiments suggest that germ cell-less and mtlrRNA are required for germ-cell formation (18, 19), while antisense experiments with pgc1 suggest a role for this gene in germ-cell migration or development (20). Other maternal factors localized to the posterior pole in an oskar-dependent manner include maternal hsp83 mRNA, whose RNA is stabilized in the posterior depending on the germ plasm; the cell cycle regulator cyclin B; outspread; and the Fat-facets protein that is a sequence-specific de-ubiquitinating proteinase. The roles of these genes in germ-cell formation, migration, or development are unclear.