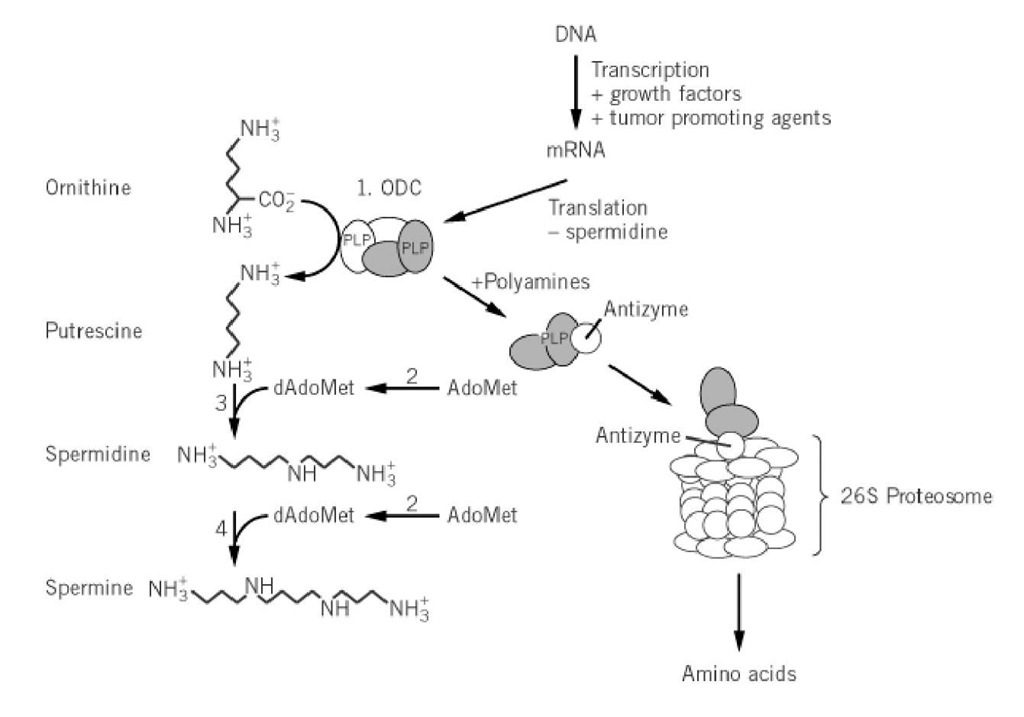
Ornithine decarboxylase (ODC; EC 4.1.1.17), first discovered in 1968, catalyzes the pyridoxal-5′ phosphate (PLP)-dependent decarboxylation of L-ornithine to generate putrescine (Fig. 1), the first committed step in the biosynthesis of polyamines (1-9). The polyamines putrescine, spermidine, and spermine are ubiquitous to all cells and are required for cell growth and differentiation. Consequently, ODC has been found in almost every cell type studied (4, 10-14). Overexpression of ODC or induction of its biosynthesis by growth stimuli in eukaryotic cells causes cell transformation . Inhibitors of ODC arrest cell growth and are actively studied for their therapeutic potential as anticancer (1, 7, 8, 15) and antimicrobial agents (2, 5). To mediate the consequences of depletion or overexpression of ODC, higher eukaryotes tightly regulate levels of the enzyme through transcriptional, translational, and posttranslational mechanisms (reviewed in Refs. 4, 9, 16-19).
Figure 1. The polyamine biosynthetic pathway. Steps 1 to 4 are catalyzed by ODC, S-adenosylmethionine decarboxylase, spermidine synthase, and spermine synthase, respectively. The mechanisms of ODC regulation in mammalian cells are depicted and include transcriptional, translational, and posttranslational control. Not shown is an alternative pathway to putrescine via arginine decarboxylase present in bacteria and plants (4).
0.1. ODC in Tumor Promotion
Deletion of the ODC gene renders yeast (12), T. brucei (20) or mammalian (21) cells dependent on added putrescine for growth, whereas overexpression of ODC activity leads to mammalian cell transformation (22). Cell transformation caused by a number of oncogenes, growth factors, and chemical carcinogens is correlated with increased ODC activity. Inhibitors of ODC activity or ODC gene transcription block oncogene-induced transformation of rat fibroblasts (22), and 12-O-tetradecanolyphorbol-13-acetate-induced formation of tumors in epidermal cells (23, 24). Overexpression of eukaryotic initiation factor eIF-4E, which increases ODC translation, also causes cell transformation, and this transformation is also blocked by ODC inhibitors or by the expression of a dominant-negative mutant of ODC (25). Tissue-specific overexpression of ODC in various transgenic mice models causes increased tumor development in skin (26, 27), reduced fertility in testis (28), and hair loss in follicles (29), although overexpression in brain has no long-term effects on tumor incidence or neuronal degeneration (30, 31). Finally, ODC activity correlates with the acquisition of hormone-independent, poorly differentiated tumors in a rat breast cancer model (32) and of a multidrug resistance phenotype (10, 11). Thus, ODC may be useful as a clinical marker.
0.2. ODC as a Drug Target
A wide array of ODC inhibitors have been designed and tested, but by far the most experimentally detailed studies have been done with mechanism-based inhibitors. They and their clinical uses have been extensively reviewed (2, 4-8, 33). The best-characterized of these inhibitors, a-difluoromethylornithine (DFMO; eflornithine), was first described by Metcalf and co-workers. In early studies in vitro and in animal models, DFMO was a promising antitumor agent, but clinical studies in humans failed to demonstrate any significant impact on tumor burden. Currently, DFMO is being investigated as a chemoprevention agent for colon and cervical cancer (15, 22, 34-37).
In contrast to the poor potency of DFMO against cancer, in 1980 Bacchi and co-workers demonstrated that DFMO cures mice infected with Trypanosoma brucei, the causative agent of African sleeping sickness. Since this landmark discovery, DFMO has been approved for treating Gambian trypanosomiasis in humans (reviewed in Ref. 2). The basis for the selective toxicity continues to be debated. Differences in the mammalian and T. brucei enzymes’ ability to bind DFMO are not involved, but several metabolic differences have been found between mammalian and T. brucei cells that are likely to contribute to the selective toxicity:
1. Mammalian ODC is rapidly degraded intracellularly, whereas T. brucei ODC is stable, suggesting that differential rates of protein degradation are a basis for selective drug toxicity ((2), (38)).
2. Trypanosomes utilize trypanothione, a conjugate of glutathione and spermidine, to maintain cellular redox balance, and this novel role of spermidine may make the cells more sensitive to polyamine depletion (39).
3. DFMO elevates the levels of S-adenosylmethionine in trypanosomes, but not in mammalian cells, and may result in building up toxic S-adenosylmethionine metabolites (40).
4. Mammalian cells possess a high-throughput transporter for putrescine, whereas T. brucei cells do not (39). In contrast, the intracellular parasitic protozoa Trypanosoma cruzi rely on scavenged polyamines to sustain growth (39).
0.3. ODC Structure/Function Analysis
The genes (or cDNA) that encode ODC have been reported from a wide variety of organisms, including many mammalian sources, fungi, parasitic protozoa, and bacteria (Prosite entries PS00878 and PS00703). Interestingly, the eukaryotic and prokaryotic enzymes do not share significant sequence identity and differ in their three-dimensional structures. Thus, ODC activity has evolved independently at least twice (41). The eukaryotic ODC are homologous to biosynthetic arginine decarboxylase from bacteria and plants, to diaminopimelate decarboxylase from bacteria (Prosite ID PS00878) and to alanine racemase (41). In contrast, prokaryotic ODC belongs to the same family as lysine decarboxylase and biodegradative arginine decarboxylase (Prosite ID PS00703). The three-dimensional structure of bacterial ODC from Lactobacillus, determined by X-ray crystallography, has structural similarity to aspartate aminotransferase (42).
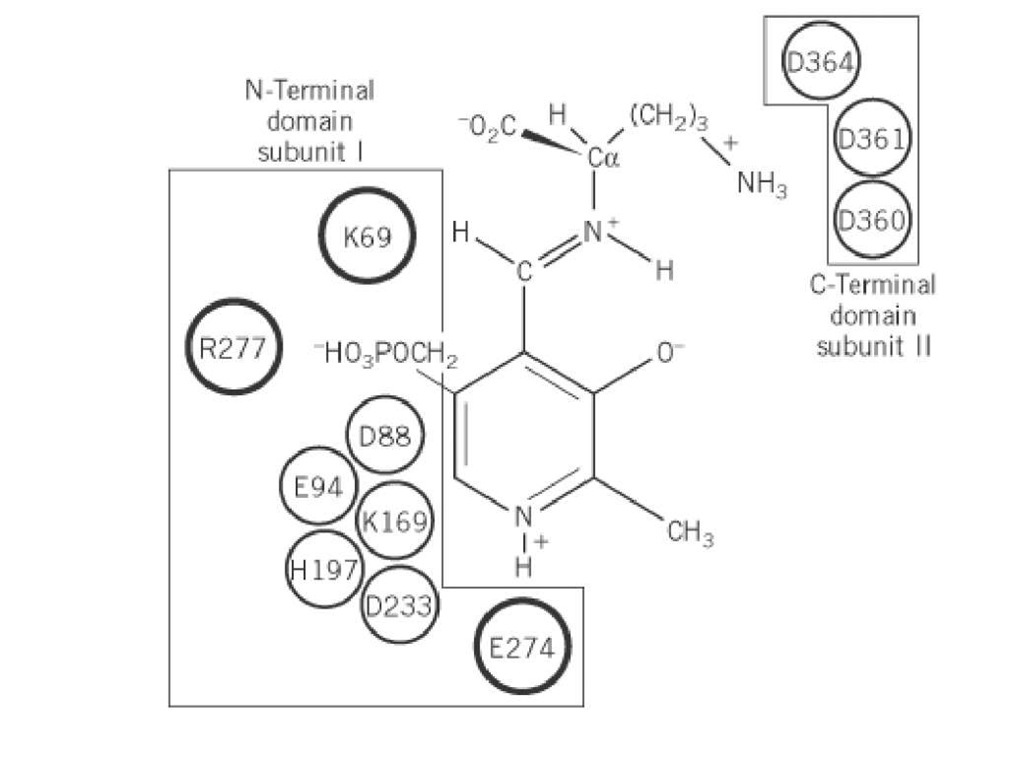
A structural model for the N-terminal domain (300 amino acids) of eukaryotic ODC predicted that it folds into a b/a barrel, and the PLP binding site formed by the C-termini of the b- strands is at the center of the barrel (41). This model places Lys69, His197, Asp233, Glu274, the Gly-rich loop (residues 235 to 237), and Arg277 in the PLP binding site and is strongly supported by the available biochemical data (discussed later). Now, both mouse (43) and T. brucei (44) ODC have been successfully crystallized, and the mouse ODC structure confirms that it folds into a b/a barrel (M. Hackert, personal communication).
ODC is an obligate homodimer, and each subunit is organized into two domains composed of a large N-terminal domain (residues 1 to 305) and a small C-terminal domain (residues 306 to 425), as demonstrated by the finding that separate polypeptides that represent these domains in T. brucei ODC can be coexpressed to yield an active tetramer that has kinetic properties identical to the native dimer (50). Mouse ODC may also be circularly permutated at amino acid 308 without significantly affecting activity (51). Two identical active sites are formed at the dimer interface and are composed of residues from the N-terminal domain of one subunit and the C-terminal domain of the other, as demonstrated by the finding that activity is restored upon mixing the inactive Lys69Ala mutant ODC with the inactive Cys360Ala ODC for the enzymes from mouse (1), T. brucei (45), or L. donovani (45). The subunits of mouse and T. brucei ODC are in rapid equilibrium, and heterodimers are formed immediately upon mixing two different homodimers. In contrast, the subunits from L. donovani ODC dissociate only after the enzyme is partially denatured in urea. Additionally, fully functional heterodimers can be formed between mouse and T. brucei ODC (45). This result suggests that the contacts in the dimer interface, which are essential for dimerization, are conserved between T. brucei and mouse ODC.
Three assays have been described to follow the decarboxylation of ornithine: a 14CO2 assay (1), a dye-linked spectrophotometry assay (45), and a circular dichroism assay (46). The kcat for the decarboxylation of L-ornithine is typically 10s-1, and the K m ranges from 90 to 400 |iM (47, 48). The enzyme is also active on lysine, but the Km is 100-fold higher than for ornithine (48). The reaction of T. brucei ODC with L-ornithine proceeds through a quinoid intermediate, and product release, not decarboxylation, was determined as the rate-limiting step (49).
The roles of a number of residues in the active site of ODC have been delineated by biochemical analysis and site-directed mutagenesis. By reduction and peptide mapping, Lys69 was identified as the residue that forms the Schiff base with PLP, and Cys360 was identified as the site of covalent attachment of DFMO (1). Consistent with these findings, mutation of both residues significantly reduces kcat (1). Mutation of Glu274 to Ala decreases kcat 50-fold, but wild-type activity is restored by substituting N-methyl-PLP for PLP in the reaction. This demonstrates that Glu274 interacts with the protonated pyridine nitrogen of PLP to enhance the electron-withdrawing capability of the ring (48). The Arg277 to Ala mutant has decreased PLP binding affinity and an altered P-NMR spectra of the enzyme-bound PLP (52), supporting the hypothesis that Arg277 interacts with the 5′ phosphate of PLP. Mutation of Asp361 to Ala increases the Km for ornithine 2000-fold and has little effect on kcat, suggesting that Asp361 forms part of the substrate-binding pocket (48). Mutation of a number of other conserved residues also results in large decreases in enzyme activity, but the roles of these residues in catalysis remain unknown (1, 48). Taken together, these results produce a picture of the ODC active site in which PLP is bound by the large N-terminal domain and interacts with Lys69, Glu274, and Arg277, whereas the C-terminal domain of the second subunit is likely to contribute to substrate binding through interactions with residues 360 to 364. These results suggest that the substrate binds across the dimer interface (Fig. 2).
Figure 2. Schematic representation of the active site of eukaryotic ODC. PLP is displayed bound to ornithine via a Schiff base. Amino acids necessary for enzyme activity by site-directed mutagenesis are displayed. The three residues that have established functions are outlined in large circles.
0.4. ODC Regulation
0.4.1. Rapid Turnover of ODC and its Polyamine-Dependent Regulation
ODC from higher eukaryotes (e.g., mammals), one of the most rapidly degraded proteins known, has an intracellular half-life ranging from 10 to 60 min (the mechanism has been reviewed in Refs. 18 and 19). Polyamines accelerate the turnover rate from the already rapid basal levels by increasing the concentration of a 26-kDa regulatory protein, termed the antizyme, which was first discovered for its ability to inhibit ODC activity. Polyamines regulate antizyme production by inducing ribosomal frame-shifting (53). The antizyme binds to the ODC monomer and targets it for ubiquitin-independent degradation by the 26 S proteosome (19, 54, 55). The antizyme is further regulated by the presence of antizyme inhibitor, a protein that is structurally related to ODC, but is missing many catalytic residues and cannot dimerize with native ODC (56). Antizymes that share similarity to the mammalian protein have also been found in other higher eukaryotes, for example, Xenopus laevis (57), where increased ODC turnover is associated with gastrulation (58). In contrast the ODC inhibitory proteins found in Escherichia coli are unrelated to the mammalian protein and are likely to play a very different role in ODC regulation (59).
Structural elements of ODC responsible for its rapid degradation have been delineated. Two PEST sequences (mouse ODC residues 298 to 333 and 423 to 461), which are associated with rapidly degraded proteins, are found in mammalian ODC (19) but not in the stable T. brucei enzyme (2). The C-terminal PEST sequence (423 to 416) is required for degradation and is sufficient to confer degradation on the stable T. brucei enzyme when expressed in mammalian cells. Mouse ODC expressed in trypanosomes is also stable, even in the presence of coexpressed rat antizyme (60), suggesting that trypanosomes lack the cellular machinery necessary to target the mouse enzyme for degradation. The requirement for the C-terminal PEST sequence for degradation in mammalian cell systems is further illustrated by the findings that mouse ODC is stabilized by truncation of as few as five residues from the C-terminus, internal deletions in the C-terminus, masking of the free C-terminus, mutation of Cys441 to Trp, or phosphorylation of ODC (reviewed in Refs. 19 and (61)). In addition to requiring the C-terminal degradation domain, polyamine-dependent degradation requires an additional part of the sequence (residues 117 to 140 in the mouse sequence), which interacts with the C-terminal half of the antizyme (reviewed in Refs. 18 and 19). The N-terminus of the antizyme is required to degrade mouse ODC and further confers lability when fused to the N-terminus of stable proteins in an in vitro assay (62, 63). Regulation of ODC in single-cell eukaryotes is less well studied than in mammalian cells. Yeast ODC is rapidly degraded by the proteosome (64). Neurospora crassa ODC is also rapidly degraded, but polyamines do not increase the degradation rate. However, in many other lower eukaryotes ODC is stable (reviewed in Ref. 19; see the previous discussion of T. brucei ODC).
0.4.2. Transcriptional regulation
Transcription of the mammalian ODC gene is increased in response to a number of growth stimuli. Basal levels of ODC transcription are mediated by several DNA elements in a complex cell type-specific manner, for example, the cAMP-responsive element CRE (65) and Sp1 (66). ODC transcription is stimulated above basal levels by protein kinase A through CRE in adrenal carcinoma cells (67), by c-Fos in PC12 cells (68), by c-Myc (69), and by the c-Myc.Max protein complex (70, 71). Overexpression of ODC in a mouse myeloma cell line (72) or in mouse myeloid 32D.3 cells induces apoptosis, and ODC is a mediator of c-Myc-induced apoptosis in the latter cells (73). The transcription factor AP-2 acts as a negative regulator of Myc-induced ODC gene transcription (74). The Wilms’ tumor suppressor represses activity of the ODC promoter and suggests that tumor suppressors may have a role in maintaining normal ODC levels (75). Rotenoids (23) and tyrosine kinase inhibitors (24) inhibit 12-0-tetradecanolyphorbol-13-acetate-induced ODC transcription and may have utility as chemoprevention agents.
0.4.3. Translational regulation
Although many factors influence the levels of ODC messenger RNA, it is well established that the levels of mRNA do not always correspond to the ODC activity observed. For example, the growth-related changes in ODC mRNA are 10-fold less than the corresponding ODC activity upon stimulation of Ehrlich ascites tumor cells. In quiescent cells high mRNA levels contrast with low protein levels. These observations suggest that ODC levels are controlled partially at the level of translation (76). It is predicted that the 5′ untranslated region (5′UTR) of the mammalian ODC gene folds into a stable secondary structure, and deletion analysis demonstrates that the 5′UTR inhibits translation of both ODC and heterologous gene products (reviewed in Ref. 1). Alternative splicing of the 5′UTR relieves the translational suppression of ODC in tumor-derived pancreatic acinar cells and results in constitutive expression (77). The polyamines spermidine and spermine inhibit ODC translation, and this inhibition depends on the presence of the 5′UTR. Insulin-induced ODC translation also depends on the 5′UTR (1, 78). ODC activity or the activity of a reporter construct linked to the 5′UTR are greater in cells that overexpress eIF-4E, a eukaryotic transcription factor that has RNA helicase activity which unwinds the secondary structure of RNA. Increased ODC expression in these cells results from changes in polyamine regulation and decreased translational repression (79, 80).
ODC is an essential enzyme that promotes cell growth and controls differentiation. Consistent with its important function, the enzyme is highly regulated in mammalian cells. The continued study of both ODC structure/function and of ODC regulation should lead to a greater understanding the role of ODC in cell function and to the design of additional methods to combat cancer and infectious disease.