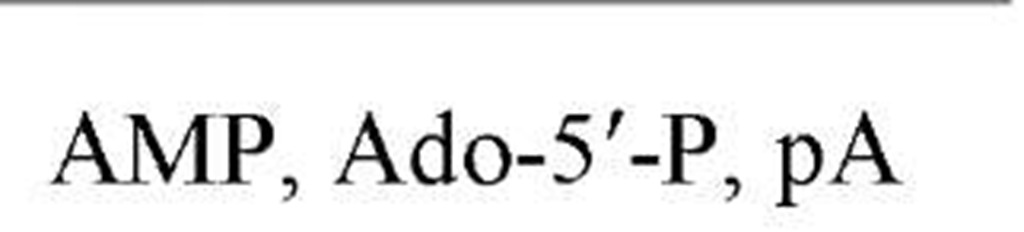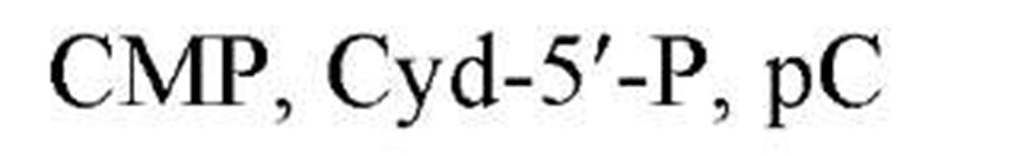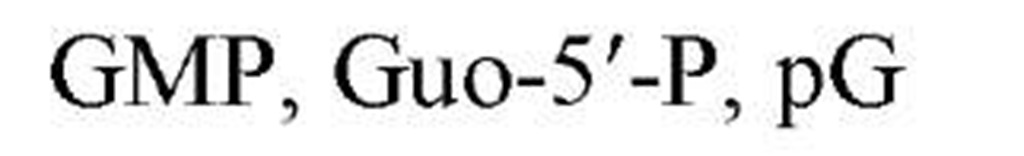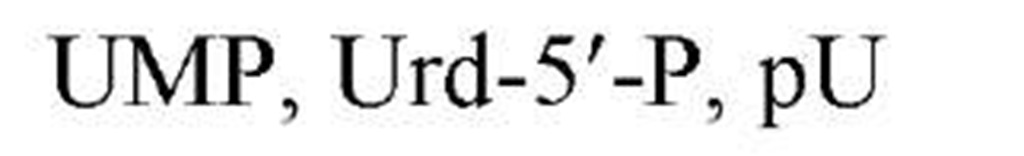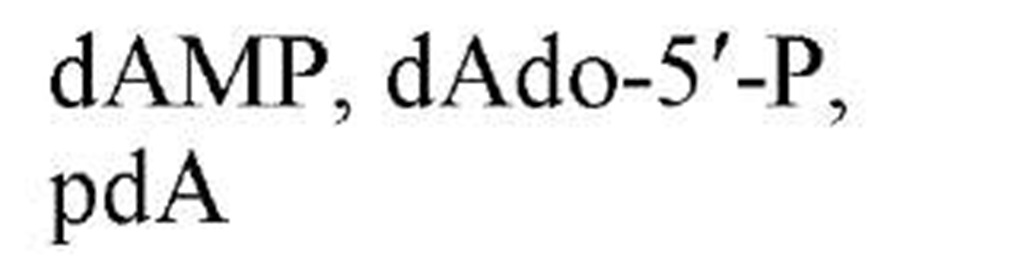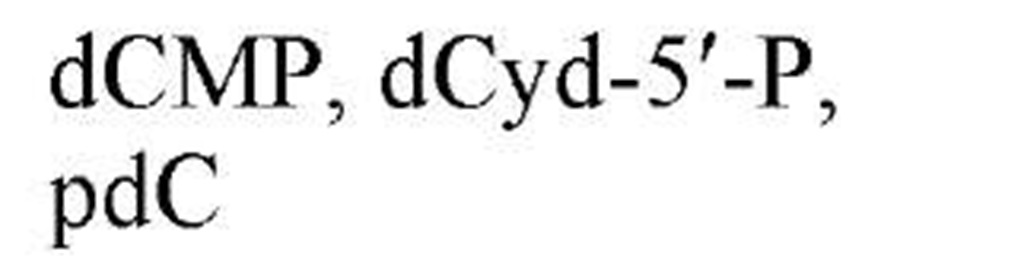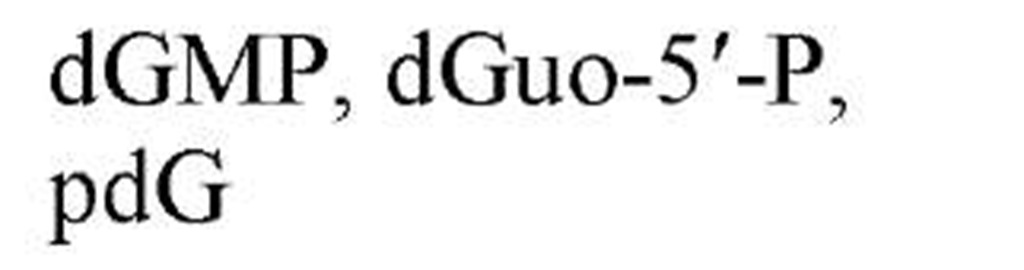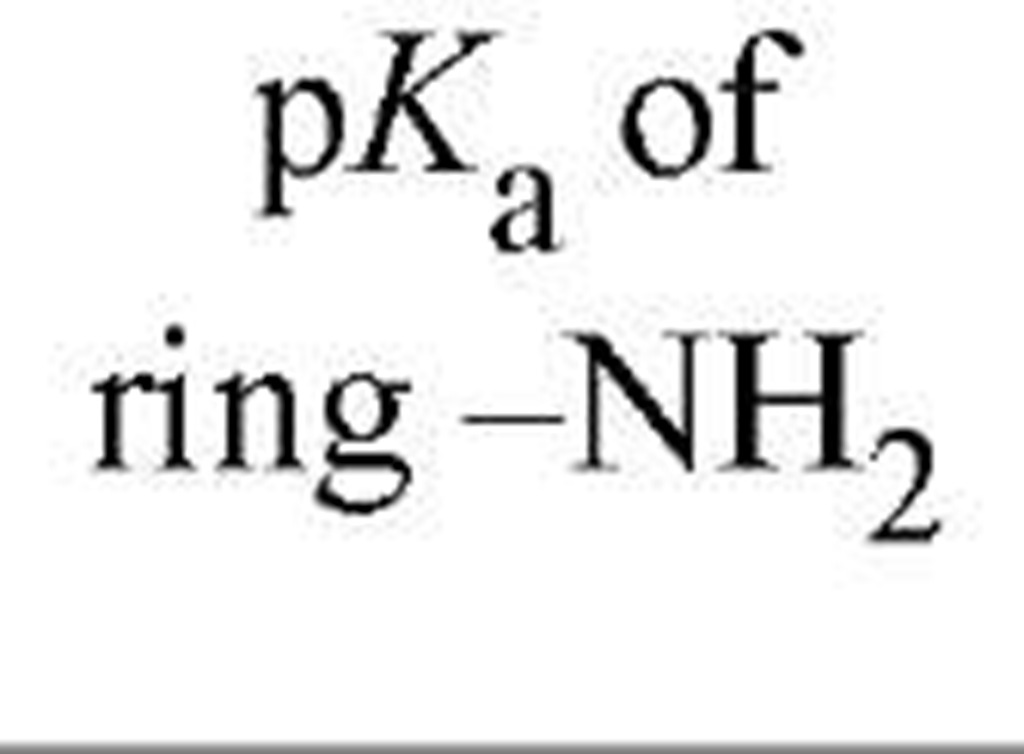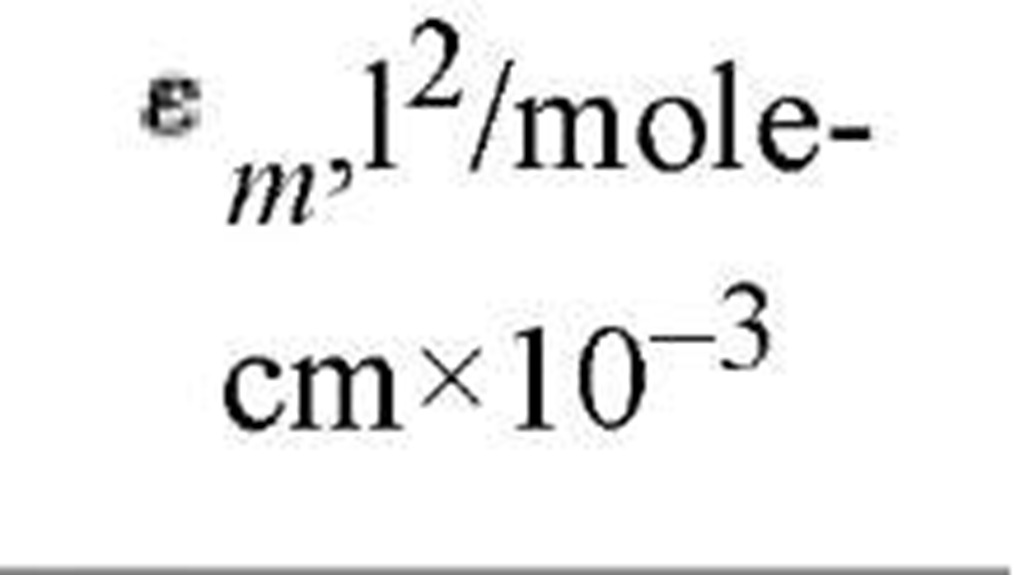Nucleotides are the monomeric constituents of nucleic acids, as well as structural elements of most coenzymes and many activated metabolic intermediates. This article describes the structure, nomenclature, and chemical and physical properties of nucleotides that serve as nucleic acid constituents.
1. Definitions
A nucleotide is a substance that, on hydrolysis, yields per mole at least 1 mole of a nitrogenous base, a sugar, and orthophosphate. A nucleoside yields 1 mole each of a nitrogenous base and a sugar. The term nucleobase refers to those nucleotides found in nucleic acids—adenine, guanine, cytosine, and uracil in RNA, and adenine, guanine, cytosine, and thymine in DNA, plus the quantitatively minor bases found in both nucleic acids. Other bases, such as the nicotinamide found in NAD+ and NADP+, occur as parts of coenzymes or as parts of metabolically activated biosynthetic intermediates, such as uridine diphosphate glucose.
A mononucleotide is a nucleotide that, on hydrolysis, yields 1 mole each of a base and a sugar plus at least 1 mole of orthophosphate. The term nucleoside diphosphate usually refers to a mononucleotide which, on hydrolysis, yields 1 mole each of a base and a sugar plus 2 moles of orthophosphate; similarly, a nucleoside triphosphate yields 3 moles of orthophosphate per mole of nucleotide. A dinucleotide yields, on complete hydrolysis, 2 moles each of base and sugar, plus at least 1 mole of orthophosphate; a trinucleotide yields 3 moles each of base and sugar, plus 2 or more moles of orthophosphate.
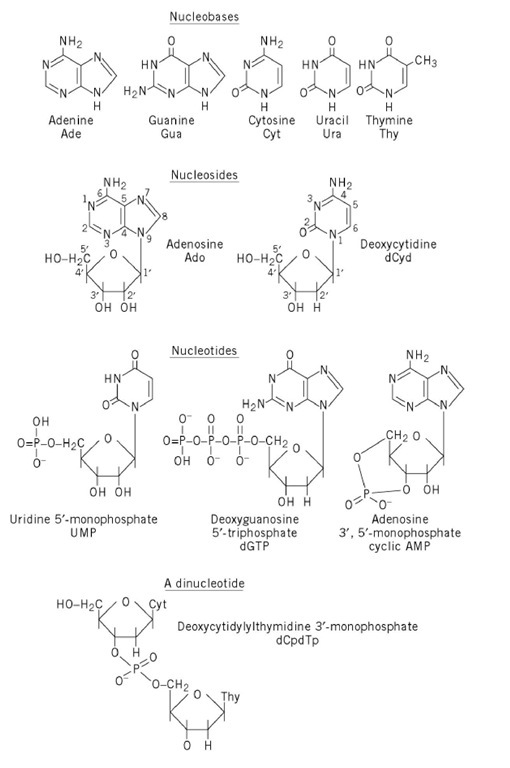
Figure 1 shows the structures of the five common nucleobases (in their most frequent tautomeric configuration) and several nucleosides, mononucleotides, and oligonucleotides, as well as the numbering systems used. Note that, in all nucleotides found in nucleic acids, a glycosidic bond links N-9 of a purine base or N-1 of a pyrimidine base to C-1, the carbonyl carbon, of either ribose or 2-deoxyribose. By convention, all positions in the sugar of a nucleoside or nucleotide are given primed numbers. Thus, the adenine nucleotide recovered from digestion of DNA with certain enzymes would be named 2′-deoxyadenosine 5′-monophosphate, identifying the position of the phosphate esterified to the sugar, as well as the carbon of the sugar that is in the reduced, or deoxy, configuration. Note that some modes of nucleic acid digestion will yield 2′- or 3′-nucleotides, in which the phosphate is esterified to positions 2′ or 3′ of the sugar, respectively. Also, in the 3′,5′-cyclic nucleotides, cyclic AMP and cyclic GMP, which play multiple roles in biological signal transduction, the phosphate is esterified simultaneously to carbons 3′ and 5′. Because the nucleic acid biosynthetic intermediates are nucleoside 5′-phosphates, or 5′-nucleotides, these structures are shown predominantly. Table 1gives the names and common abbreviations of the nucleotide constituents of nucleic acids and the cyclic nucleotides.
Figure 1. Structures of the five common nucleobases and representative nucleosides and nucleotides.
Table 1. Names and Abbreviations of the Nucleotide Constituents of Nucleic Acids and of Cyclic Nucleotides
a Thymine occurs as a minor base constituent of tRNAs. However, because the thymine ribonucleotides do not occur in nature, nucleotides containing thymine are called either thymidine or deoxythymidine nucleotides. No consistent preference for either nomenclature has emerged.
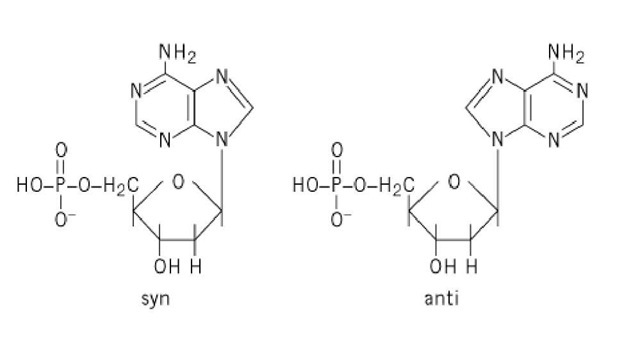
One additional structural feature of nucleotides relates to the somewhat hindered rotation about the glycosidic bond, which leads to two rather stable orientations of bases with respect to the sugar, termed syn and anti (Fig. 2). Because it is more compact to draw, most of the structures shown here are represented as syn, but the anti configuration predominates in nucleic acids (see Z-DNA).
Figure 2. dAMP in the syn and anti configurations.
2. Chemical and Physical Properties of Nucleotides
Because of the phosphate group or groups on nucleotides, these substances are highly acidic and exist as anions at physiological pH. The primary and secondary p^a values for the phosphate groups of nucleoside 5′-monophosphates are about 1 and 6, respectively. The amino groups on adenine, guanine, and cytosine rings are extremely weak bases, undergoing protonation with p^a values of 2 to 4.5. Because of variations in these values, and the absence of a ring amino group in uracil and thymine (see Table 2 and ref. 2), one can adjust the pH to values at which different nucleotides have different net negative charges, thereby permitting their ready separation by electrophoresis or ion-exchange chromatography (2).
Table 2. Properties of Nucleoside Monophosphates
a N.R., not reported.
b Data for cyclic nucleotides are from Ref. 10. Other data are from Ref. 1.
The aromatic character of the purine and pyrimidine rings leads the nucleotides to absorb ultraviolet light with characteristic absorption spectra, as indicated in Table 2. This allows ready detection, identification, and quantification of nucleotides and nucleotide derivatives. Because of ionization of the purine and pyrimidine rings, the ultraviolet absorption spectra are quite dependent on pH, and this has made identification and quantification methods even more specific and sensitive.
3. Naturally Occurring Modified Nucleotides
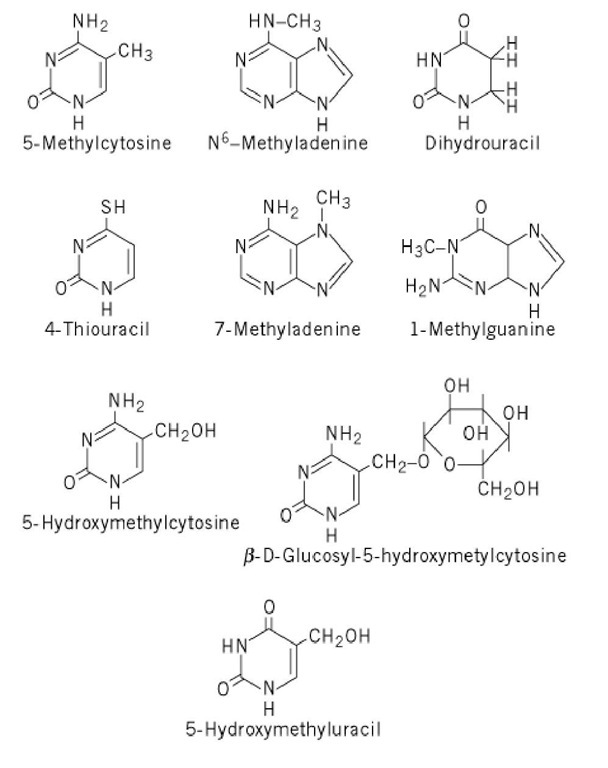
In addition to the five "standard" nucleobases (adenine, guanine, cytosine, thymine, and uracil), most nucleic acids contain modified bases (see Transfer RNAand DNA Structure ). The most widespread is 5-methylcytosine, which is present at up to 5% of the cytosine level in most eukaryotic DNA (Fig. 3). The most abundant methylated base in prokaryotic DNA is N6-methyladenine (see Methyltransferase, and Mismatch Repair). A variety of modified bases, some but not all of them methylated, are found in RNA, most abundantly in transfer RNA. A few of these are shown in Figure 3.
Figure 3. Structures of prominent modified nucleobases found in nucleic acids.
In addition to the above modified bases, which are quantitatively minor in most systems studied, there are several bacteriophages whose DNA contains modified bases as major constituents. Most prominent among these are the T-even coliphages, which contain four forms of 5-hydroxymethylcytosine, completely substituted for cytosine (Fig. 3): the base itself, with no further modifications, and the base glycosylated at the hydroxymethyl position with glucose in either the a or b configuration, or with cellobiose (glucosyl-1,4-b-D-glucose). Several phages infecting Bacillus subtilis contain 5-hydroxymethyluracil completely replacing thymine, and bacteriophage XP-12, which infects Xanthomonas oryzae, contains 5-methylcytosine completely replacing cytosine. 5-Hydroxymethyluracil has also been found as a minor constituent of DNA subjected to oxidative damage; it results from oxidation of thymine residues (3).
4. Synthetic Nucleoside and Base Analogues
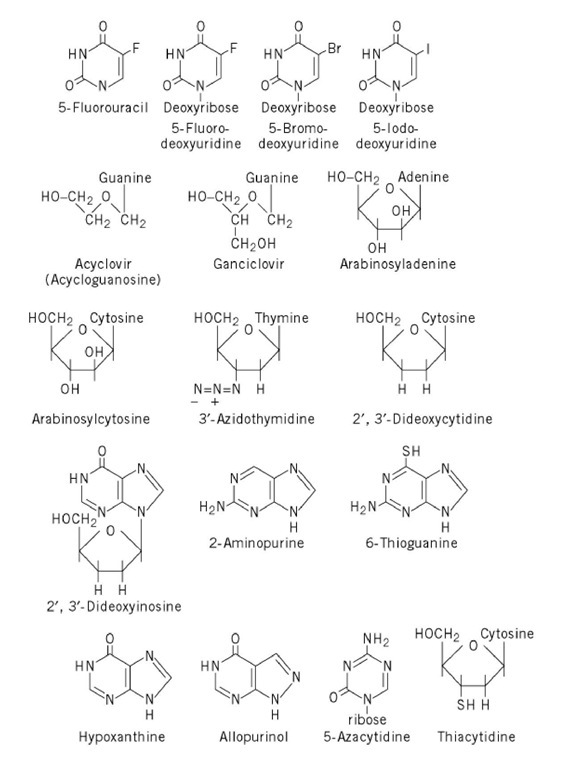
A large number of nucleoside and nucleobase analogues have been synthesized and developed as anticancer, antiviral, antibacterial, and antiparasitic drugs (4-6). Many of these analogues act by intracellular conversion to deoxyribonucleotide analogues, which selectively block DNA replication (because deoxyribonucleotides have no significant metabolic roles other than as DNA constituents). A few of the most prominent of these analogues are shown in Figure 4 and discussed briefly here.
Figure 4. Nucleobase and nucleoside analogues of biological or medical significance.
4.1. Halogenated Pyrimidines
The earliest-developed useful synthetic nucleobase and nucleoside analogues were 5-fluorouracil (FUra) and 5-fluorodeoxyuridine (FdUrd), both of which were synthesized during the 1950s and shown to have potent antitumor activity; both agents are still in use today. Both the base and the nucleoside act by uptake into target cells, followed by intracellular conversion to 5-fluorodeoxyuridine 5′-monophosphate (FdUMP), an analogue of deoxyuridine monophosphate, which binds covalently to thymidylate synthase, thereby blocking the synthesis of dTTP and hence the replication of DNA.
Because of their negative charge, nucleotides are not readily transported through cell membranes. With most nucleobase or nucleoside drugs, the active form of the drug is a nucleotide analogue— FdUMP in the example under discussion. Thus, in order to use nucleic acid precursors successfully as drugs, the biochemical pharmacologist must be aware of the nucleotide biosynthetic salvage enzymes in target cells (see Salvage Pathways To Nucleotide Biosynthesis), so that a precursor is administered that can be converted to the active nucleotide. One must be concerned also with degradative enzymes. For example, nucleoside phosphorylase can cleave fluorodeoxyuridine to 5-fluorouracil plus deoxyribose-1-phosphate, a reaction that would limit the effectiveness of the drug by decreasing its availability for conversion to the active FdUMP.
Closely related to 5-fluorodeoxyuridine are the other halogenated pyrimidines, 5-bromodeoxyuridine (BrdUrd) and 5-iododeoxyuridine (IdUrd). The bromine atom of BrdUrd has a van der Waals radius similar to that of a methyl group; thus, it is readily phosphorylated by thymidine kinase and ultimately converted to the nucleoside triphosphate BrdUTP, which is a close dTTP analogue and is readily incorporated into DNA. BrdUrd has limited therapeutic utility, but it is widely used as a research reagent, for density-labeling experiments involving DNA; the heavy bromine atom imparts a significantly higher buoyant density to BrdUrd-substituted DNA than to thymidine-containing DNA. Moreover, BrdUrd is a potent chemical mutagen because bromouracil in DNA occasionally base-pairs with guanine and not with thymine in DNA ( (6); also, see Mutagenesis). By contrast, IdUrd is a fairly useful antiviral drug, used in treating eye infections caused by the large DNA virus, herpes simplex. The genome of this virus encodes a thymidine kinase of broad specificity that readily phosphorylates IdUrd to the monophosphate, and then to the diphosphate. Thus, this nucleotide is more readily incorporated into DNA of virus-infected cells, where its presence can damage the viral genome, than in uninfected cells.
4.2. Arabinosyl Nucleosides
These compounds, of which the adenosine and cytidine analogues are most useful therapeutically (6), contain arabinose, which is the 2-epimer of ribose. AraA is a fairly effective antiviral agent, being used to treat viral encephalitis, among other conditions. The triphosphate of araA, araATP, is a rather specific inhibitor of DNA polymerases encoded by herpes viruses. AraC is used in chemotherapy of leukemias and some solid tumors. AraC is phosphorylated initially by deoxycytidine kinase, ultimately being converted to araCTP, a potent inhibitor of replicative DNA polymerases. A problem in the use of araC is its fairly facile deamination by cytidine deaminase, yielding the weakly effective arabinosyluracil. Therapeutic activity of araC can be potentiated by coadministration of a cytidine deaminase inhibitor. In the same sense, the efficacy of araA is often enhanced by coadministration of an adenosine deaminase inhibitor, because this enzyme converts arabinosyladenine to arabinosylhypoxanthine.
4.3. 6-Thioguanine
This guanine analogue is widely used in somatic cell genetics. Its conversion to a toxic nucleotide is carried out by hypoxanthine guanine phosphoribosyltransferase (HGPRT). Since the structural gene for HGPRT is sex-linked, mammalian genomes contain only one copy of the gene, making this gene a convenient selectable marker for studies on mutagenesis and for generating hybridoma cell lines for monoclonal antibody production. HGPRT-negative mammalian cell mutants become resistant to the lethal effects of 6-thioguanine, because of their inability to anabolize the analogue.
4.4. Allopurinol
This hypoxanthine analogue is a successful drug for treating gout (7). It is an effective inhibitor of xanthine oxidase, which converts hypoxanthine to xanthine, and xanthine to uric acid in the catabolism of purine nucleotides. Thus, allopurinol often inhibits the buildup of the insoluble uric acid, allowing excess purines to be excreted as the more soluble hypoxanthine and xanthine.
4.5. 5-Azacytidine
This cytidine analogue has a nitrogen atom at position 5 of the modified pyrimidine ring. It has been useful in studies of the biological significance of DNA methylation because it is metabolized similarly to cytidine. When incorporated into DNA as the dCMP analogue, however, the pyrimidine ring cannot be methylated at position 5. Thus, a G-mC base pair is effectively demethylated in subsequent rounds of replication, allowing investigators to investigate the epigenetic changes that occur as a result.