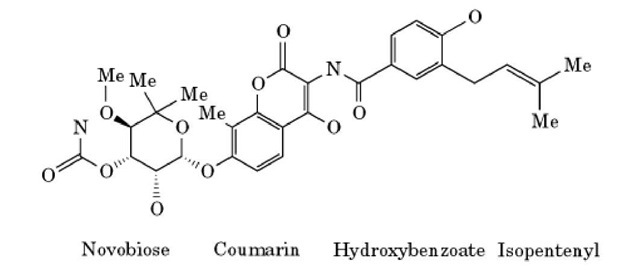
Novobiocin is an antibiotic of the aminocoumarin class that was originally discovered in 1955 (1). It is produced by bacteria of the species Streptomyces spheroides and Streptomyces niveus (2). Novobiocin is composed of a noviose sugar linked to a substituted coumarin and a prenylated 4-hydroxybenzoic acid (Fig. 1). Because of its pronounced side effects, novobiocin is not the antibacterial treatment of choice in the clinic (3). Nevertheless, it has regained some interest recently due to its synergistic effect with some antitumor drugs (4, 5).
Figure 1. The chemical structure of novobiocin.
The antibacterial activity of novobiocin is based on its inhibition of bacterial type-II DNA topoisomerases (see DNA Topology). The type-II DNA topoisomerases are essential cellular enzymes that, together with the type-I DNA topoisomerases, determine DNA superhelicity and are involved in chromosomal condensation and segregation (6, 7). Using the energy derived from ATP hydrolysis, type-II topoisomerases introduce a double-stranded cut into a duplex DNA, pass a second duplex through the opening, and then reseal the cut. In prokaryotes, the type-II topoisomerases DNA gyrase and DNA topoisomerase IV are both tetramers of two subunits (8). The A subunit of DNA gyrase (ParC in topoisomerase IV) harbors the DNA breakage-reunion activity, and the B subunit of DNA gyrase (ParE in topoisomerase IV) is responsible for ATP hydrolysis.
The bacterial-type II topoisomerases are the targets of several antibiotic compounds. Quinolones interact with the A subunit, coumarins (eg, novobiocin) and cyclothialidines (9) with the B subunit. Novobiocin (10, 11) and the cyclothialidines (12) block the ATPase activity of the B subunit of DNA gyrase by binding to the ATP-binding site. The detailed binding mode of novobiocin and of an ATP analog to a 24-kDa N-terminal fragment of the Escherichia coli DNA gyrase B protein has recently been determined by X-ray crystallography (13). The structure shows that ATP and novobiocin have partially overlapping binding sites. Therefore, novobiocin is a competitive inhibitor of ATP binding to the gyrase B subunit. An extensive hydrogen-bonding network is formed between novobiocin and the enzyme. Most of the bonds are contributed by the novobiose sugar group that forms hydrogen bonds with Asn46, Ala47, and Asp73. In addition there are several hydrogen bonds to ordered water molecules, which in turn hydrogen bond to Val43, Val71, Asp73, Gly77, and Thr165. The coumarin ring of novobiocin forms two hydrogen bonds to Arg136, and it is also involved in hydrophobic interactions with Arg76 and Pro79. The prenylated hydroxybenzoate moiety of novobiocin is not required for DNA gyrase inhibition (14) and shows few interactions with the enzyme. This group is important, however, for the antibacterial activity of novobiocin, possibly by influencing uptake of the compound by bacterial cells. The crystal structure of chlorobiocin, a close homologue of novobiocin, bound to the 24-kDa N-terminal fragment of the gyrase B subunit is also known (15). This information is very helpful in understanding differences in the affinity of various novobiocin derivatives, and it could be exploited in rational drug design.
The occurrence of resistance to novobiocin is a frequent phenomenon. Mutation of the gyrase B subunit gene that leads to an altered gyrase B subunit is the most widespread mechanism of novobiocin resistance. A summary of the known mutations leading to novobiocin resistance is given in Table 1. It is obvious that the conserved residue Arg136 (E. coli numbering) is most often involved in resistance development. This residue has also been mutated in two coumermycin A-resistant strains of Borrelia burgdorferii (16). The susceptibility of these strains to novobiocin has not been tested, but coumermycin-resistant strains are normally cross-resistant to novobiocin. Arg136 forms two hydrogen bonds with the coumarin ring in the crystal structure of the 24-kDa N-terminal fragment of E.coli gyrase B with novobiocin. Therefore, involvement of this residue in resistance development is not surprising (13). The effects of mutation of Arg136 on the structure of the 24-kDa N-terminal fragment and on the binding affinity and the binding mode of novobiocin have recently been described (17). Mutation of Arg136 decreases the ATPase and supercoiling activity of the DNA gyrase B subunit (18), but the residual activity of the enzyme is still sufficient to fulfill the essential task of DNA gyrase. Novobiocin-resistant mutants of gyrase B must still retain sufficient activity to carry out the essential role of DNA gyrase, so it is not surprising that the mutated residues are not directly involved in binding ATP but instead surround the ATP-binding site.
Table 1. Mutations in the gyrB Gene from Novobiocin-Resistant Strains
|
Organism |
Mutation of Gyrase Ba |
Increase in MIC- or IC50- |
|
Escherichia coli |
R136L |
>8x |
|
R136C |
12x |
|
|
R136S |
13.2x |
|
R136H |
11.6x |
|
|
G164V |
>3x |
|
|
Staphylococcus aureus |
G85S (G77) |
32x |
|
I102S (I94) |
32x |
|
|
S128L (V120) |
8x |
|
|
R144I (R136) |
64x |
|
|
I102V, T173N |
32x |
|
|
(T165) |
32x |
|
|
S128L, A108S |
64x |
|
|
(A100) |
250x |
|
|
R144I, I175T |
||
|
(V167) |
||
|
R144I, I102S |
||
|
Streptococcus |
S127L (V120) |
128x |
|
pneumoniae |
||
|
Haloferax |
D82G (G81), S122T (S121), R137H (R136) |
No data |
a The numbers in brackets refer to the residue in E. coli homologous to the mutated residue in the respective species. The amino acid residues are indicated by their one-letter abbreviations. b MIC: Minimal inhibitory concentration; IC5Q: 50% inhibitory concentration.
In addition, gyrase B resistance to novobiocin can also result from an active process of efflux from the cell. Recent results have demonstrated that the baseline level of resistance of Haemophilus influenzae and Pseudomonas aeruginosa to novobiocin is caused by multidrug efflux pumps that actively extrude novobiocin (19, 20). The novobiocin-producing organism Streptomyces sphaeroides protects itself against the toxic effects of novobiocin by producing two gyrase B subunits, one of which is resistant to the drug (21). Two loci that confer novobiocin resistance have been identified in Streptomyces niveus (22). Both loci also hybridize to DNA from S. sphaeroides but they do not encode a gyrase because they do not hybridize to the DNA gyrase gene.
Novobiocin is widely used in basic research, to study the effects of DNA topology on gene expression. DNA topology is influenced by many parameters, such as osmolarity and temperature. Novobiocin is used to mimic the consequences of such parameters on DNA topology. By exposing cells to novobiocin, it was demonstrated that DNA relaxation leads to increased expression of the sigma factor that governs the heat-shock response and to a concomitant induction of the production of heat-shock proteins (23, 24). Similarly, it was shown that DNA supercoiling also regulates the expression of genes that respond to osmotic pressure (25). Furthermore, the inhibition of DNA gyrase by novobiocin leads to an increase in the expression of the genes that encode the enzyme (26).