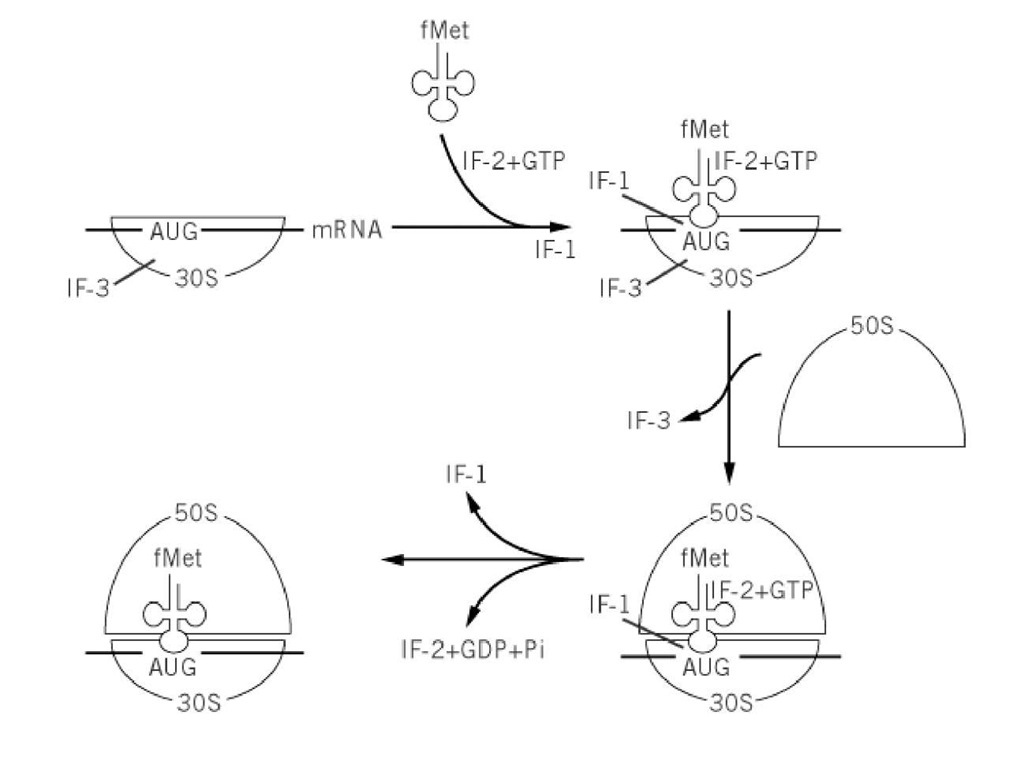
Formation of an initiation complex for translation of protein biosynthesis on 70 S ribosomes in bacteria involves, in addition to initiator fMet-tRNAfMet, messenger RNA, and GTP, the cooperative interaction of three proteins, initiation factors IF-1, IF-2 and IF-3 (Table 1). Until now, no convincing data have indicated the existence of additional factors. Numerous in vitro experiments demonstrate that IF-1 and IF-3 shift the ribosome-dissociation equilibrium from 70 S ribosomes toward dissociation into 30 S and 50 S subunits. Under physiological conditions, the ribosomal subunit equilibrium is toward 70 S formation, and IF-1 serves to increase the rate constant for both the dissociation and association of 70 S ribosome, whereas IF-3 shifts the equilibrium strongly toward dissociation by binding to the 30 S particle. IF-3 also selects the initiator tRNA in the presence of the initiation codon triplet, as well as on a natural mRNA template. IF-3 action distinguishes the initiator transfer RNA from elongator tRNAs by specific domains in the tRNA and not by its formyl methionine (fMet). Part of the information is located in the anticodon stem and loop region. IF-3 has been shown to destabilize selectively polynucleotide-primed 30 S complexes formed with elongator tRNAs. The third initiation factor, IF-2, directs fMet-tRNAfMet binding to the 30 S subunit. This initiation factor might also exclude noninitiator tRNAs from the P site, while allowing formylated charged initiator tRNA to enter. This reaction is stimulated by IF-1 and GTP. In addition, IF-2 helps the association of the two subunits and has a GTPase activity in the presence of ribosomes. The process of initiation complex formation in prokaryotes is summarized in Fig. 1 (1).
Figure 1. The initiation pathway of protein synthesis in prokaryotes.
Table 1. Escherichia coli Initiation Factors
|
Factor |
Molecular Mass |
Number of Amino Acids |
Gene |
Function |
|
IF-1 |
8,119 |
71 |
infA |
Promotes IF-2 and IF-3 functions |
|
IF-2a |
97,300 |
889 |
infB |
Binds fMet-tRNA and GTPase |
|
IF-2b |
79,700 |
732 |
infB |
|
|
IF-3 |
20,668 |
181 |
infC |
Ribosome dissociation and mRNA binding; increasesthe specificity of the initiation complex for fMet-tRNA |
In the absence of tRNA or initiation factors, the 30 S particle by itself binds to the proper initiation region on mRNAs. The sequence of events directed by initiation factors shows that IF-1 and IF-3 are released before or during the joining of the subunits, whereas IF-2 is released after their joining and requires GTP hydrolysis. The three factors are present in cells at sufficiently high concentration to saturate the free 30 S particle. The steady-state levels of initiation factors are coordinately regulated in exponentially growing bacteria relative to one another and relative to ribosome levels. All levels rise as a function of increasing growth rate, suggesting that the cellular levels of initiation factors are under metabolic control. Neither IF-2 and IF-3 synthesis is under stringent control in vivo.
All of the initiation factor genes have been cloned and mapped: infA for IF-1, infB for IF-2, and infC for IF-3. They are dispersed on the bacterial chromosome and, with the exception of infA., are associated with other components of the protein biosynthesis apparatus (threonyl and phenylalanyl-tRNA synthetase, and ribosomal protein L20 and L35 for IF-3; tRNAfMet, NusA, and ribosomal protein S15 for IF-2). The infB gene produces two forms of IF-2, IF-2a (97.3 kDa), and IF-2b (79.7 kDa), using two in-frame initiator codons. Both forms are implicated in fMet-tRNA binding to 30 S ribosomes and possess a ribosome-dependent GTPase activity. The meaning of the existence of two forms of IF-2 in bacteria, as well as their functional difference, is not known.
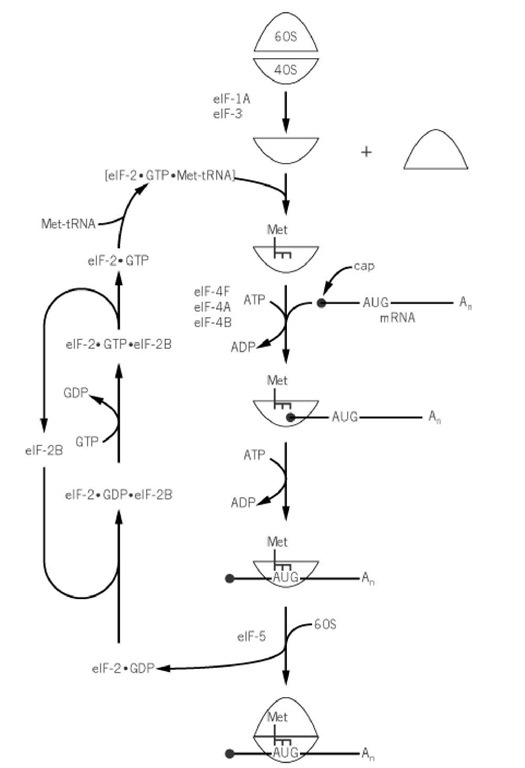
The eukaryotic initiation steps are promoted by at least 10 different initiation factors, some of which are themselves complexes of numerous protein subunits. Figure 2 summarizes where the more important of the initiation factors function in the pathway (2). eIF-1A and eIF-3 are involved in the dissociation of ribosomes; eIF-2 and eIF-2B are primarily concerned with tRNAlMet binding to 40 S subunits; eIF-4A, eIF-4B, and eIF-4F promote mRNA binding and scanning; and eIF-5A is involved in the junction step. Essentially all the initiation factor polypeptide cDNAs have been cloned and sequenced (Table 2).
Figure 2. The initiation pathway of protein synthesis in eukaryotes.
n pathway of protein synthesis in eukaryotes.
Table 2. Eukaryotic Initiation Factors (2)
|
Factor |
Subunit Molecular Mass (kDa) |
Function |
|
eIF-1 |
12.6 |
Enhances initiation complex formation |
|
eIF- |
16.5 |
Ribosomal dissociation; promotes Met- |
|
1A |
tRNAiMet binding |
|
eIF-2 |
125 |
Binds Met-tRNAiMetand GTP |
|
|
a |
36.1 |
Phosphorylated subunit |
|
|
b |
38.4 |
Binds Met-tRNAiMet |
|
|
g |
51.8 |
Binds GTP; Met-tRNAiMet |
|
|
eIF- |
270 |
Guanine nucleotide exchange factor for |
|
|
2B |
a |
33.7 |
eIF-2 |
|
b |
39.0 |
Binds GTP |
|
|
g |
58 |
Binds ATP |
|
|
d |
57.1 |
Binds ATP |
|
|
e |
80.2 |
Phosphorylated subunit |
|
|
eIF- |
94 |
Stabilizes ternary complex in presence of |
|
|
2C |
RNA |
||
|
eIF-3 |
650 |
Dissociates ribosomes; promotes Met-tRNAiMet and mRNA binding |
|
|
p35 |
35 |
Highly phosphorylated |
|
|
p36 |
36.5 |
||
|
p40 |
39.9 |
||
|
p44 |
35.4 |
RNA binding motif |
|
|
p47 |
47 |
||
|
p48 |
52.5 |
Identical to Int-6 (oncogene) homologue |
|
|
p66 |
64.0 |
Binds RNA |
|
|
p110 |
105.3 |
||
|
p116 |
98.9 |
Major phosphorylated subunit |
|
|
p170 |
166.5 |
||
|
eIF- |
44.4 |
ATPase, helicase, binds RNA |
|
|
4A |
|||
|
eIF- |
69.8 |
Binds RNA, promotes helicase activity |
|
|
4B |
|||
|
eIF-4F |
250 |
Binds 5′ Cap, helicase |
|
|
eIF-4E |
25.1 |
Cap-binding subunit |
|
|
eIF-4A |
44.4 |
ATPase, helicase |
|
|
eIF-4G |
153.4 |
Binds eIF-4A, eIF-4E, eIF-3, and Pab1p |
|
|
eIF-5 |
48.9 |
Promotes GTPase with eIF-2 and ejection of eIFs |
|
|
eIF-6 |
25 |
eIF-2 forms a ternary complex with Met-tRNAlMet and GTP that binds to the 40 S ribosome, where it is established by eIF-1A and eIF-3. The resulting 43 S preinitiation complex then interacts with mRNA prepared by eIF-4F, eIF-4A, and eIF-4B. eIF-4F is a heterodimeric complex that binds to the -Cap structure of mRNA. It is composed of one molecule each of eIF-4E, eIF-4A, and eIF-4G. eIF-4E is of lower abundance in the cell than the other subunits and initiation factors, thereby making the eIF-4F complex rate-limiting under most circumstances. eIF-4A is an RNA-dependent ATPaseand binds RNA only weakly. Its sequences contain ATP-binding elements found in the DEAD box family of RNA Helicases. eIF-4B is an RNA-binding protein that contains two RNA-binding motifs: the RNP consensus octamer and hexamer elements and an arginine-rich domain. eIF-4F (bound to the 5′ Cap) together with eIF-4B possess RNA helicase activity dependent on ATP hydrolysis. When mRNA secondary structure is melted, the 40 S ribosome is able to bind to the 5′ Cap region and begin scanning. Proper interaction of the Met-tRNA iMet anticodon with the AUG initiator codon is monitored by eIF-5, which promotes GTP hydrolysis, followed by the ejection of initiation factors and junction of the 40 S initiation complex with 60 S subunits. eIF-2 leaves as a complex with GDP and requires eIF-2B-catalyzed exchange of the GDP for GTP in order to promote another round of initiation.