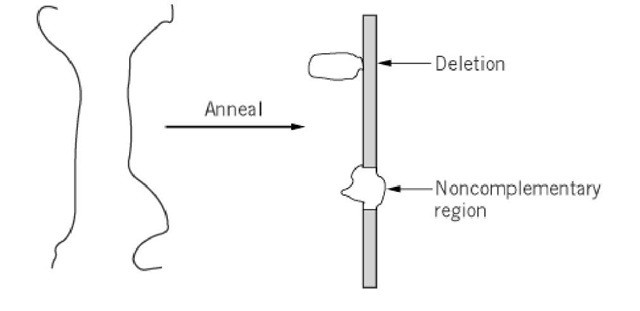
Two single-stranded nucleic acid polymers with at least partial complementarity can form duplexes with stretches of double helix, corresponding to the regions of nucleotide sequence homology, interleaved with unpaired stretches, corresponding to noncomplementary regions. Two types of single-stranded loops are introduced by noncomplementarity in the heteroduplex (Fig. 1). A loop in only one of the strands indicates a deletion. A loop composed of single-strand regions in both strands indicates a region of sequence incompatibility. A heteroduplex can be composed of two DNA strands, one DNA strand and one RNA strand, or two RNA strands. In vivo, heteroduplexes (DNA-DNA) can arise in the process of recombination or as a consequence of errors (mismatches) in DNA replication (DNA-DNA) or transcription (DNA-RNA). Hybridization of nucleic acids of different origins leads to heteroduplexes in vitro.
Figure 1. Loops in a heteroduplex indicate either a deletion or a region of noncomplementarity in the two strands.
The level of sequence homology in the heteroduplex can be evaluated by monitoring the change in ultraviolet absorbance on heteroduplex formation or by selective enzymatic digestion of single-stranded regions, followed by hydroxyapatite chromatography. These techniques are analogous to those used for studies of reassociation kinetics (1).
Electron microscopy can used to visualize the regions of sequence homology in the heteroduplex, even though nucleic acids are not sufficiently dense to be easily observable directly in the electron microscope. The heteroduplex is spread on the surface of the electron microscope grid. Heavy metals and proteins are applied to the nucleic acid by one of several techniques, thereby making the polymers visible in the electron microscope.