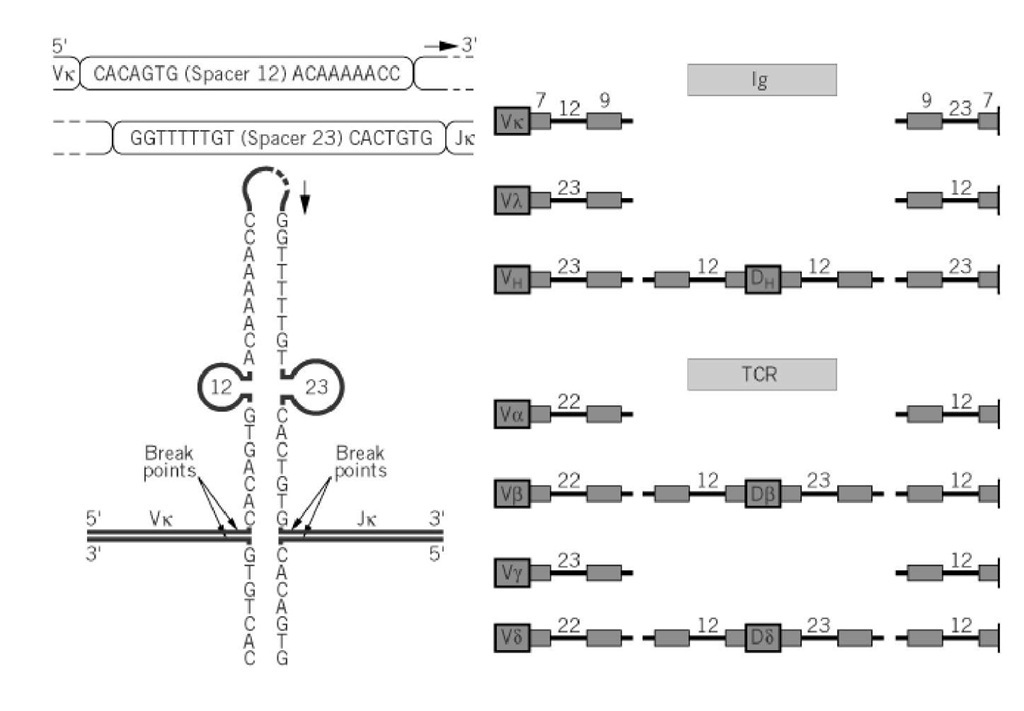
The unique feature of immunoglobulin gene organization is that these genes need to be rearranged before becoming functional. These rearrangements allow the immune system to generate a very large repertoire by recombination and random association of a limited number of gene segments (ie, between 200 and 300 in humans), leading to at least 10 million discrete immunoglobulins having different specificities. In the light chains, the variable region results from the random association of VL (Vk or Vl) and JL (Jk or Jl) genes. The heavy chains are more complex, because their diversity results from the random association of VH, D, and JH genes. Immunoglobulin genes are rearranged exclusively in the B-cell lineage, and these events constitute the hallmark of the B-cell differentiation in the bone marrow. Similarly, genes for the T-cell receptor (TCR) rearrange exclusively in T cells by a very similar process. Ig gene rearrangements require precise signals on the DNA, known as recombination signal sequences or (RSSs), that flank each of the V, D, and J gene segments and are recognized by highly specific recombinases encoded by two genes, termed RAG1 and RAG2 (for recombinase activating genes). RSSs consist of two palindromic sequences that are highly conserved, although not identical, between the different loci; one is a heptamer and one is a nonamer, separated by a spacer region 12 or 23 nucleotides long (Fig. 1). The two RSSs that participate to a given joint have always spacers of different lengths.
Figure 1. Recombination signal sequences of Ig and TCR genes. ( a) The 3′ end of a Vk gene is followed by an RSS tha complementary to RSS 5′ of the Jk gene onto which Vk will be connected by the recombinase. (b) Schematic organizatio of the RSS in Ig and TCR genes.
The first recombination event, DH to JH, takes place in the early proB cells and is rapidly followed by the rearrangement of one of the VH segments to D-J. Another enzyme, terminal deoxynucleotidyl-transferase, is also active in proB cells and adds nucleotides in a random fashion during the joining of D to J and of V to D-J rearrangements. Addition of these non-germline-encoded nucleotides is known as N-diversity and considerably amplifies the repertoire, because both the sequence and length of VH and VL complementarity-determining region 3 (CDR3) are modified by this mechanism. Rearrangements are strictly regulated by their end product—that is, the chain they encode once they become functional. Thus, once a m chain is synthesized, it will block the rearrangement of the second IGVH allele. This requires that the m chain be expressed as a membrane protein (mm). In a mouse rendered transgenic for a membrane VDJ-Cm chain gene, endogenous rearrangements at the IGVH locus are blocked, so that B cells will make only the transgenic m. When the same experiment is performed with a VDJ-Cm gene without the M exons (thereby encoding only the soluble form of the m chain), endogenous rearrangements are no longer blocked. The fact that m regulates rearrangement only when expressed as mm strongly suggests that it operates at the cell surface. In fact, mm is coexpressed with the so-called surrogate light chain, composed of the product of two genes, l5 and VpreB, and expressed specifically in proB and preB cells (see B Cell). l5 is highly homologous to l and is disulfide-bonded to the m chain by its penultimate cysteine residue, whereas VpreB resembles a V Ig domain. The surrogate light chain (YL) has been shown to contribute efficiently to the regulation of allelic exclusion. The negative feedback that prevents the second allele from rearranging is probably a consequence of the turning off of the terminal deoxynucleotidyl-transferase and RAG genes. The RAG genes, but not the transferase, are reactivated as the preB cells cycle and induce light-chain gene rearrangement, with the same hierarchy of control on the second allele whenever the first has rearranged. The IgK locus is activated first. If both k alleles fail to produce a functional rearrangement, the IGL enters the game, with the same type of control. Finally it should be observed that the overall process of rearrangement is not very efficient, with only about 10% final success. This is obviously the price to pay for the complexity and partial randomness of the system.
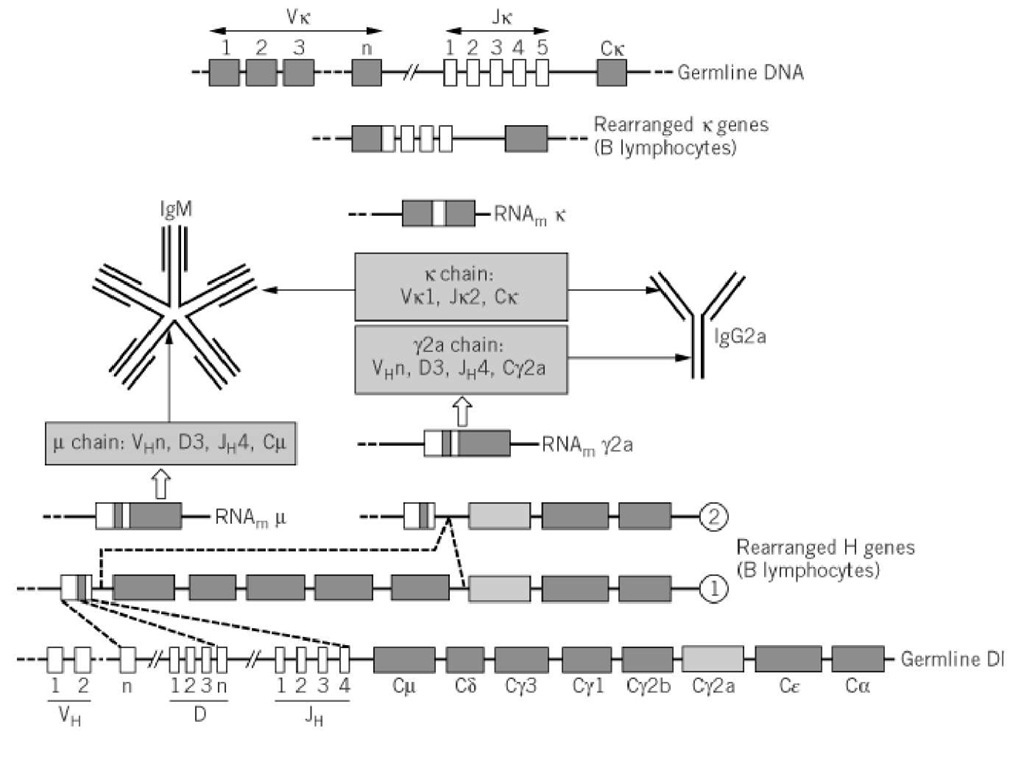
Once both H and L genes have successfully rearranged, the resulting immunoglobulin is expressed at the B-cell surface. When stimulated by an antigen, specific clones proliferate and colonize the secondary lymphoid organs (lymph nodes, tonsils, spleen, etc.), where they become organized in germinal centers, in close contact with other partners of the immune response (ie, dendritic cells and T cells). Contact with T helper (Th) cells will promote the last step of gene rearrangement, called isotype switching, by which the VDJ gene region will be associated with a new constant heavy-chain gene. The classical most frequent switch that occurs in immune responses is IgM ^ IgG. A schematic example of all the rearrangements that lead to the production of an IgG molecule is given in Figure 2.
Figure 2. Rearrangement events that lead from Ig germline genes to synthesis of IgM and IgG molecules in the mouse model. Recombination first occurs at the IGVH locus, to generate a V(D)J combination (here chosen as VHn-D3-JH4) that is transcribed with Cm and processed as a m mRNA, resulting in the expression of a m chain that will associate with k chain (here Vk1-Jk2-Ck) that results from the second wave of rearrangement targeted at the IGVK locus. IgM is first expressed as a monomer at the cell surface of immature B cell (not shown). After antigenic stimulation, the pentameric form of IgM antibody is secreted by plasma cells and is rapidly replaced by an IgG (here IgG2a), which occurs by class switching (second line from bottom). Note that the switch will not change the VDJ region, so the antibody specificity is maintained.