Prior to the development of the electron microscope, the size of an object, such as a virus, was estimated by filtration through membranes of various pore sizes. By this criterion, the bacterial virus (bacteriophage) fX174, first described in 1935 (1), and its close relative S13 (2) were classified as much smaller than the T phages that were under investigation in the early days of molecular biology. Moreover, the fact that their genetic material is DNA made these two viruses attractive candidates as front runners in the race to unravel the molecular basis of heredity. The contributions began with the initial characterization of the fX174 DNA as being single stranded, as reported in the first issue of the Journal of Molecular Biology (3). The culmination came when fX174 was the first organism to have its genome subjected completely to DNA sequencing (4), which offered the first direct documentation of the existence of overlapping genes in nature.
1. Virus structure
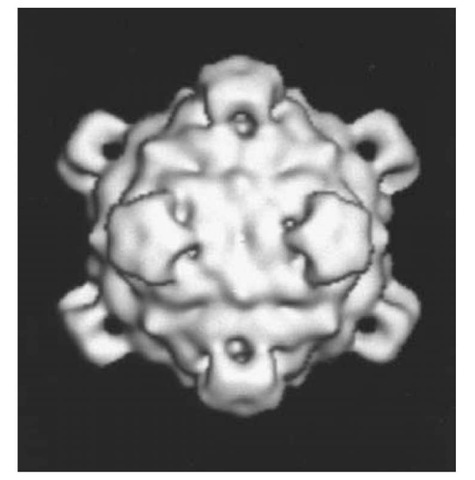
The fX174 virion appeared polyhedral with "knobs" or "spikes" in early electron micrographs of metal-shadowed and stained preparations (4). These images led to the proposal that the virion has icosahedral symmetry, a structural feature that was later confirmed at a higher resolution by cryoelectron microscopy (5) and at atomic resolution by X-ray crystallography (6). The major contribution of these structural studies was, however, the establishment of the three-dimensional protein structures of the three major proteins in the crystalline form of the fX174 virion and their quaternary structure in both the crystalline and aqueous states of the virion. In the virion, the fX174 circular, single-stranded, DNA genome is enclosed within a T = 1 icosahedral protein shell, the simplest of the possible deltahedrons (7). Consequently, the protein shell of the virion contains 60 copies of the proteins encoded in genes F, G, and J, and 12 copies of the gene H protein product (Table 1). Five copies of F capsid protein are clustered around each of the 12 vertices of the icosahedron, forming a shell that has no openings at the center of the 20 triangular faces, as seen in the reconstructed image of the virion exterior in Figure 1. Five copies of G protein form the spikes, or projections, that extend outward from the capsid surface at each vertex of the icosahedron (Fig. 1). The spikes are attached to the capsid outer surface by each G protein interacting with its F protein partner via eight direct hydrogen bonds and five indirect, water-mediated hydrogen bonds (8).
Figure 1. Three-dimensional reconstructed image of fX174 virion from electron micrographs of unstained frozen-hydrated preparations (courtesy of Norman H. Olson and Timothy S. Baker, Purdue University). The diameter of the virion is 33.5 nm between the exterior edges of the spikes at the fivefold axes (28).
Table 1. fX174 Structural Proteins
|
Protein |
Mol. Wt. |
No. of Residues |
No. in Virion |
No. in Procapsid |
|
B protein |
13.8×103 |
120a |
0 |
60 |
|
D protein |
16.9×103 |
152 |
0 |
240 |
|
F protein |
48.4×103 |
426 |
60 |
60 |
|
G protein |
19.0×103 |
175 |
60 |
60 |
|
H protein |
34.4×103 |
328a |
12 |
12 |
|
J protein |
4.2×103 |
36 |
60 |
0 |
a Based on DNA sequence (4).
The single-particle reconstruction (5) and crystallographic data (6) also revealed structural details about the interiors of the capsid layer and the spikes, as well as additional structural components that are interacting with the interior surface of the capsid. For example, both F and G polypeptides contain an eight-stranded antiparallel beta-sheet motif (b-barrel) like that found in most viral capsid proteins. In addition, each F pentameric cluster contains a small, central channel that leads into a central channel within each G pentameric cluster. This continuous channel has mainly hydrophobic walls and varies between 0.6 and 2.6 nm in diameter (6). The unassigned electron density within the G channel near the junction with the F channel is thought to be the location of a short segment of the H protein that presumably lies within the channel. The remainder of the H polypeptide chain is believed to be located beneath the interior opening of the channel, in contact with both the DNA and the interior surface of the F capsid proteins. The main evidence for this location for each of the 12 H proteins (9) is the fact that urea treatment of virions removes both the G spikes and the H proteins, leaving a spherical DNA-containing particle (10). Although the H polypeptide chains could not be unambiguously located in the X-ray crystallography electron density map, all 36 residues of the basic J protein could be assigned to an S-shaped path on the inner surface of the capsid, linking two neighboring F proteins across each two-fold axis of symmetry. In addition, a four-nucleotide segment of single-stranded DNA was located in the difference Fourier map calculated from the differences in the diffraction data on 114 S virions and on 70 S particles that contain less DNA (1). These nucleotides are interacting with four amino-acid side chains of the C-terminal segment of the J protein and with the side chains of seven amino acid residues in the F protein.
2. Genome structure
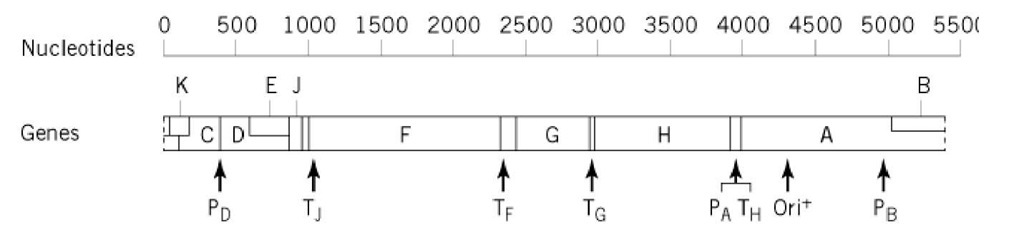
Mutational analysis has identified 11 genes in the circular genome of fX174 (Fig. 2). The A segment of the DNA sequence codes for two gene products, endonuclease A and a truncated version of A that suppresses host DNA synthesis, A (not shown in Fig. 2). The A protein is translated in the same reading frame as A, but its translation start site is 173 codons downstream of the A translation start site. Furthermore, a portion of the DNA sequence in the A segment is read in a different reading frame as gene B, one of the scaffolding proteins required for the assembly of empty procapsids (Table 1). In fact, all three reading frames are used in the DNA sequence that encodes gene K, an unidentified protein that has an effect on progeny virion yield. The protein encoded by gene C is involved in switching DNA synthesis from replication of the double-stranded fX174 replicative form (RF) DNA to the simultaneous synthesis and packaging of fX174 single-stranded progeny DNA into empty procapsids. These packaged DNA strands have the same sequence as fX174 messenger RNAs that are transcribed from the fX RF DNA. Therefore, the packaged strands are labeled positive (+), while the complementary, unpackaged strands are labeled negative (-). The next region of the genome codes for a second set of overlapping genes read in two different reading frames. D is a second scaffolding protein found in procapsids, but not in virions (Table 1), and E induces cell lysis at the bacterial site for elongation and division. Genes J, F, G, and H code for structural proteins found in both virions and procapsids (Table 1).
Figure 2. Linear version of the fX174 circular genetic map. The locations of promoters (P), transcription terminations sites (T), and the origin of replication (or/’+)) are shown.
The fX174 genome contains three types of DNA sequences, in addition to those that code for proteins and those that are involved in transcription and translation (promoters, terminators, and ribosome binding sites; Fig. 2). The first is an origin of replication (ori+) located within gene A, where endonuclease A cleaves between nucleotides 4305 and 4306 (11). The second is an "incompatibility" or "reduction" sequence (12) that interferes with fX174 replication if the sequence is present on a plasmid in the uninfected cell. The 85-nucleotide sequence includes parts of genes H and A and the intergenic sequence between them. One suggestion for the function of this sequence is the binding of the incoming single-stranded fX DNA to a site on the bacterial membrane that is essential for fX DNA replication. Presumably, these sites are present in limiting amounts in the uninfected cell. Finally, the third type of DNA sequence includes the intergenic sequences. The H/A untranslated sequence found within the incompatibility sequence shares one feature with two of the three remaining intergenic sequences (J/F, F/G, and G/H): namely, the potential for forming complementary, double-stranded hairpin structures in the single-stranded states of either the DNA or mRNA. However, these potential nucleic acid secondary structures appear to play only subtle roles in fX174 infection, since disruption of the J/F potential hairpin increased the decay rate of only two species of fX mRNA (13).
3. Infection cycle
As with most viruses, the fX174 infection cycle begins with the attachment of the virion to receptor molecules on the outer surface of the bacterial host. The core oligosaccharide region of the lipopolysaccharide (LPS) in the outer membrane of Escherichia coli C constitutes the initial sites where the fX174 virion binds via several of its spikes (14). Since O antigenic side chains attached to the core monosaccharide units block fX attachment, most gram-negative bacterial species are resistant to fX174. Binding is mediated by Ca cations (15), and there are three separate sites for the cation in the crystal structure of the virion, two in F protein near the threefold axes and one near the outer opening of the channel through the G pentamers in each spike (16). Of interest is the presence of a glucose molecule just below one of the Ca sites in F. Moreover, another host gene, which lies between gal and aroG at 17 min in the E. coli map, is required for fX binding (17).
Due to the single-strandedness of its DNA genome, fX174 has an additional functional requirement that determines which bacterial species it can infect. The virion must penetrate the outer wall and cytoplasmic membrane to deliver the fX genome to the bacterial DNA replicative complex for synthesis of the complementary (-) strand. This transfer of the viral genome marks the beginning of the eclipse phase, the time period when infectious virions are not present in the infected cell. After binding to the LPS receptor, Ca -induced structural changes in the fX virion proteins begin the ejection of the single-stranded, circular DNA from within the virion, presumably through an opening in its protein shell (18-20). Most evidence supports a model for ejection through the channel down the center of one "spike" (18, 21). However, a conformational change that would produce an opening where six F proteins meet at the threefold axis cannot be ruled out. Furthermore, it is not clear how the fX virion encounters the bacterial components involved in DNA replication. Some experiments suggest that these host factors are located where the outer wall adheres to the cytoplasmic membrane (22, 23).
As the ejected fX174 genome enters the cell, the single-stranded DNA is converted to double-stranded RF (Stage I). The host single-stranded binding protein (SSB) coats the incoming DNA, and a protein complex (the preprimosome) is assembled on the coated DNA. The preprimosome contains the sequence recognition protein (PriA), PriB, PriC, dnaT, and gyrase (dnaB) (24), and synthesis of several RNA primers begins when primase (dnaG) joins the complex. Then DNA polymerase III adds deoxyribonucleotides to the primers; DNA polymerase I replaces the RNA primers with DNA; and DNA Ligase joins the two ends of the – strand to produce a supercoiled RF molecule.
The bacterial transcription system begins to transcribe the fX174 genome, using the RF molecule as the template. Since only two fX genes (A and A ;) code for enzymes, and the remaining fX genes code for proteins that function as structural components, the virus does not require a sophisticated mechanism for regulating transcription. Except for genes E and K, the fX genes are transcribed and translated at all times during the viral replication cycle. Moreover, the relative amounts of the fX proteins are determined by the relative amounts of the mRNAs that contain the message for each protein (25), Table 2. Thus, the message for endonuclease A (and A ;) is present only in the unstable mRNA and the two least abundant transcripts (8 and 9 in Table 2), while the message for the most abundant fX protein, D scaffold (see Table 1), is found in all transcripts (Table 2). Also, the r^o-independent termination is inefficient, producing two groups of transcripts that begin at the same promoter but end at different termination sites (1, 3, 7 and 2, 4, 5, 6 in Table 2).
Table 2. fX174 mRNA Transcripts(25)
|
Genes Transcribed |
Promoter |
Termination |
Order of Abundance |
|
D, J |
PD |
TJ |
1 |
|
B, C, D, J |
PB |
TJ |
2 |
|
D, J, F |
PD |
TF |
3 |
|
B, C, D, J, F |
PB |
TF |
4 |
|
B, C, D, J, F, G |
PB |
TG |
5 |
|
B, C, D, J, F, G, H |
PB |
TH |
6, 7 |
|
D, J, F, G, H |
PD |
TH |
6, 7 |
|
F, G, H |
a PF |
TJ |
8 |
|
A, B, C, D, J |
|||
|
D, J, F, G, H |
PD |
TJ |
9a |
|
A, B, C, D, J |
|||
|
A,? |
PA |
? |
Unstable |
a Rare promoter b Single copy
Endonuclease A is the first fX protein synthesized, to create a pool of RF molecules as templates for both transcription and fX genomic DNA replication. This Stage II RF DNA replication begins with endonuclease A cleaving the + strand within the A gene sequence and forming a covalent bond with the 5 ‘ phosphate of the cleaved strand. The bacterial DNA helicase (Rep) unwinds the double-stranded RF, and DNA polymerase III adds deoxyribonucleotides to the 3′ OH of the cleaved + strand, until the new + strand is completed. At the same time, SSB coats the newly synthesized single-stranded fX DNA (26). As soon as the primosome assembly site in the F/G intergenic region (Fig. 2) of the new + strand becomes available, the preprimosome assembles on the new + strand and begins synthesis of a new – strand (see discussion of Stage I). To ensure continuous synthesis of RF, endonuclease A cleaves the + strand at the junction between the old and new strands and joins the ends of the old strand as soon as the A gene sequence on the new + strand becomes available. At the same time, A protein forms a covalent bond with the free 5′ phosphate group of the new + strand, to begin synthesis of the next RF molecule. Each new RF molecule is released from the replication complex when DNA ligase joins the ends of the completed – strand.
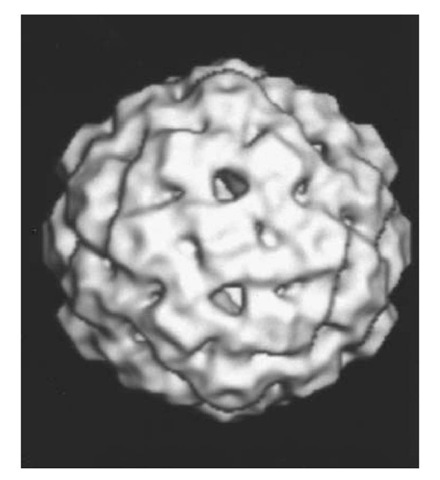
As the pool of RF molecules increases, the level of fX transcripts reaches a point where sufficient amounts of fX capsid (F), spike (G, H), and scaffold (B, D) proteins are available to begin the assembly of 108 S empty procapsids (27). First, 9S complexes are formed of F and 6S complexes of G (both containing five copies of the corresponding fX proteins). These two complexes are joined with H and assembled into a procapsid, with B crosslinking F pentamers on their interior surface across the twofold axes and D forming an exterior scaffold on the surface of the capsid (28, 29). In Figure 3, the pentameric G spikes at each fivefold axis are not as prominent as in the virion (Fig. 1) due to interactions with D on the capsid outer surface. These G/D interactions keep each G b-barrel attached to the capsid surface, but inclined at a 20° angle to the fivefold axis. In the virion, the G b-barrels are roughly parallel to the fivefold axis and held to the capsid surface with the aid of bound water molecules. In the procapsid, the mainly alpha-helical D scaffold protein is present in four radically different environments. However, protein-protein interactions between D molecules form a scaffold that holds each F and G pentameric complex in a shell that has almost three times the internal volume as the protein shell of the mature fX virion. Furthermore, the enlarged procapsid shell has holes with a diameter of 3.0 nm at the threefold axes (see Fig. 3).
Figure 3. Three-dimensional reconstructed image of fX174 procapsid from electron micrographs of unstained frozen-hydrated preparations (courtesy of Norman H. Olson and Timothy S. Baker, Purdue University). The diameter of the procapsid is 35.5 nm between the exterior edges of the spikes at the fivefold axes (28).
As procapsids are being assembled, fX C protein begins to compete with SSB for binding to RF at the fX A/helicase complex on the replicating fX DNA. Once bound, C stops Stage II RF replication and provides a binding site for procapsid, presumably at one of the 3.0-nm holes. fX + strand is then simultaneously synthesized and packaged, as 60 molecules of the basic J polypeptide are bound to the single-stranded DNA (30, 31). Each J protein displaces a B polypeptide on the interior surface of two neighboring F proteins as the single-stranded DNA is packaged. The 60 B chains presumably exit from the procapsid through the 19 other holes, forming the 132S provirion.
Meanwhile, small amounts of fX lysis (E) protein are slowly being translated from mRNA containing both D and E messages. The low level of E translation is probably due to a weak ribosome binding site, to rare codons in the E reading frame, and to competition from ribosomes translating D messages. fX E polypeptides begin forming a tunnel through the cell wall at the site of cell elongation and division (32). Once the cytoplasmic membrane extrudes through the tunnel, lysis occurs, and the fX provirions are exposed to the ionic environment of the medium. The D scaffold proteins dissociate from the provirions, allowing conformational changes in F capsid proteins and minor changes in inclination of G proteins in the spikes, as the protein shell collapses around the fX DNA. The newly formed progeny fX virions then begin the infection cycle when they encounter uninfected cells.
4. Related bacteriophage
Although most of the experimental data have been obtained with fX174, the Seventh Report by the International Committee on the Taxonomy of Viruses includes 38 other phages in the Microviridae family. The majority of the members belong to the same genus ( Microvirus) as fX. The natural hosts for the members of the Microvirus genus are various species of Enterobacteriaceae. There is, however, at least one member in three additional genuses of the Microviridae family that infect other eubacterial species. These include members of gram-negative genuses Bdellovibria and Chlamydia and the wall-less genus Spiroplasma, which causes several plant diseases.