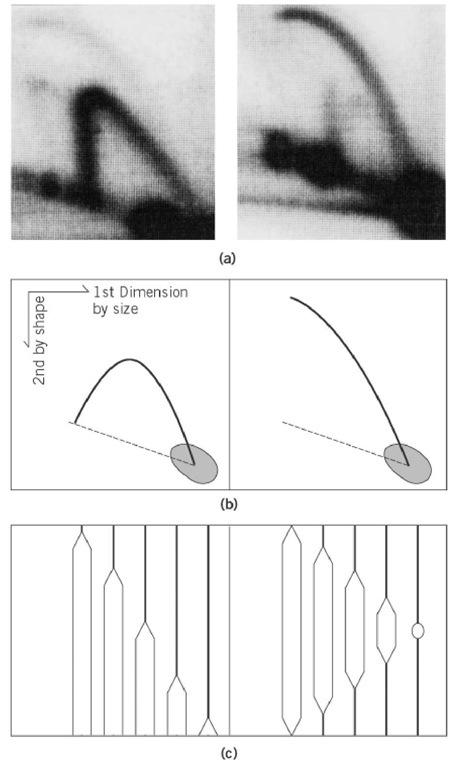
In general, DNA replication of a chromosome starts from a genetically fixed site and proceeds bidirectionally. At the initial stage of replication, the replicated regions form structures like an eye (or a bubble). This form is important, as it represents the origin region of the chromosome (see Replication Origin); various methods to detect eye-form intermediates have been developed. Electron microscopy (EM) of the replicating DNA can detect eye-forms as large as several kilobases (1). Using a combination of digestion by appropriate restriction enzymes and electron microscopy, the origin of replications of plasmids, bacteriophage, and virus DNA has been identified (2). Recently, the two-dimensional gel electrophoresis method was developed to detect various forms of replication intermediates (3). Chromosomal fragments produced by digestion with restriction enzyme are separated by electrophoresis by agarose gel electrophoresis, first by size and then by shape. The distribution of intermediates on the two-dimensional (2D) gel is detected by hybridization with a radiolabeled probe of a cloned DNA fragment from or near the replication origin. Since the correlation between the size and complexity of shape of eye-forms is different from that of Y-fork intermediates (see Replication Fork (Y-Fork Intermediate)), the former can be distinguished from the latter by the shape of arcs on the 2-dimensional gel (Fig. 1). In particular, when the origin of replication is not located at the center of the fragment, the change from eye- to Y-form occurs as replication proceeds on the fragment, which results in a discontinuous distribution of arc from eye- to Y-form. This method has been used successfully to detect the chromosomal origins of eukaryotes, in particular of Saccharomyces cerevisiae (4), Schizosaccharomycespombe (5), and Drosophila chromosomes (6). Detection of eye-forms from mammalian chromosomes is technically difficult, due to the large amount of background DNA, and this method generally produces ambiguous results as to the site of origin of replication (7).
Figure 1. Two-dimensional gel electrophoresis method to detect eye-form (bubble-form) and Y-form replication intermediates. (a) Examples of patterns of Y-form (left) and Eye-form (right) intermediates of the replication of Saccharomyces cerevisiae chromosome are shown. Intermediates were detected by hybridization with the 32P-labeled probe of cloned DNA within the fragments shown in (c). (b) The photos in ( a) are drawn schematically. Dotted lines indicate the location of linear DNA fragments of corresponding size. (c) The population of the intermediates detected in (a) and (b) Left panel: Replication fork moves from the left (or right) side of the fragment and proceeds to produce Y-forks of variable size. Theoretically, a Y-fork with equal branch lengths is structurally most complex and shows least mobility in the second dimension. Right panel: Replication initiates from the center of the fragment and proceeds bidirectionally, producing various sizes of bubble intermediates. The largest bubble is like a circular DNA and shows least mobility in the second dimensions. The locations of the probe used for detection in (a) are indicated by dotted lines. Dark spots in the photos in (a), and drawn schematically in (b), indicate that the majority of DNA detected by the probe is nonreplicated (or completely replicated) linear molecules.