Expression systems are used for the production of protein from recombinant DNA molecules (see Recombinant Proteins). They are of widespread use in industry, health care, and scientific research because of their flexibility and ability to achieve production levels exceeding those of the native source of the protein. An expression system consists of a vector carrying the gene encoding the protein of interest, along with the sequences necessary for transcription of the DNA into messenger RNA and translation of the mRNA into protein, plus a host providing the enzymatic machinery for carrying out these processes. In a homologous expression system, the gene to be expressed derives from the same species as the host whereas, in a heterologous expression system, the gene to be expressed and the host are of different origin.
1. Design of the expression vectorg
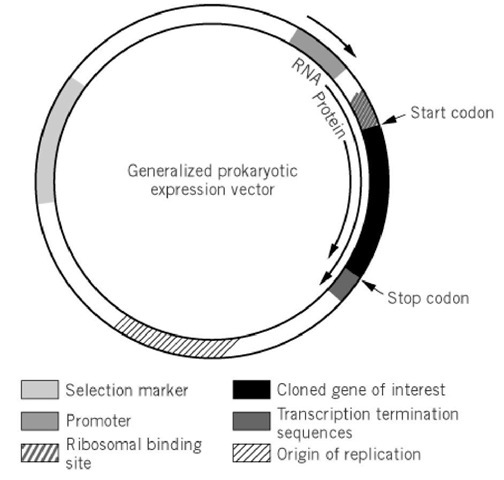
An expression vector not only provides the components necessary for cloning, transfer, stability, and multiplication of recombinant DNA but it also delivers the elements required for correct transcription of DNA into functional mRNA and efficient translation of mRNA into the desired protein. However, the importance of the basic elements controlling gene dosage and selection should not be ignored in the design of an expression system. A generalized prokaryotic expression vector is shown in Figure 1.
Figure 1. Generalized prokaryotic expression vector. The figure shows a typical prokaryotic expression vector, with a selection marker gene allowing stable maintenance of the plasmid, an origin of replication controlling the copy number of the vector, and a promoter and a ribosomal binding site positioned upstream of the gene of interest. One or more transcriptional termination sequences are positioned downstream. Furthermore, a eukaryotic expression vector would contain an extra origin of replication and an extra selectable marker to allow replication and maintenance in both E. coli and the eukaryotic host. The transcriptional termination signal would include a polyadenylation signal.
A promoter is a region of DNA recognized by an RNA polymerase and is the prerequisite for initiation of transcription. Promoters consist of characteristic sequence elements. In general, promoters from prokaryotes and eukaryotes differ, and a common organization pattern can be given for each. However, there can be pronounced differences between species, and more subtle variations are found within the same species, giving rise to promoters with different strengths and control mechanisms.
The strength of a promoter determines the frequency with which a gene is transcribed by controlling the rate at which the RNA polymerase and the promoter form the initiation complex. However, the strength is governed not only by the interaction with the RNA polymerase. Other proteins, so-called transcriptional activators and repressors, bind to the promoter region and regulate the process of transcriptional activation. Often, eukaryotic expression vectors provide transcriptional enhancer elements as well.
The ideal promoter for the expression of a recombinant protein not only generates the synthesis of high levels of mRNA but, even more important, it should be inducible, ie, the experimenter should be able to control its activity. However, only a few inducible promoters are tightly controlled, so that some transcription occurs even in the uninduced state. The overproduction of a protein will most often influence the cell in a negative manner and can be detrimental in the most severe cases, where the protein of interest is toxic to the host (see Poison Sequence). If this seems to be a problem, it may prove advantageous to use a weaker promoter under tighter control. Also, when overexpression seems to outstrip the host’s post-translational modification, molecular chaperone, and proofreading systems, the use of a weaker promoter may be beneficial. For most expression systems, both constitutive and inducible promoters are available. The "on" state of the best inducible promoters can be regulated, thereby providing a means of optimization (1).
Some prokaryotic expression vectors contain antitermination elements, which stabilize the RNA polymerase on the DNA template to ensure optimal elongation of the transcript. Transcriptional terminators positioned at the 3′-end of the expressed gene restrict the size of the mRNA, minimize the sequestering of RNA polymerase, and isolate the plasmid’s replication functions, thereby stabilizing the plasmid. Eukaryotic mRNAs are processed more extensively than prokaryotic mRNA, including such processes as splicing and the addition of poly-A tails. The signals for these steps are provided by eukaryotic expression vectors at positions downstream of the cloned gene.
The degradation of mRNA allows a biological system to adapt to changes in the environment (see RNA Degradation In Vitro). Yet the aim of the gene expression systems is to maintain a high level of mRNA for the gene of interest during its expression. The stability of mRNA can, in some cases, be increased by insertion of sequences that fold into stable secondary structures at the ends of the mRNA, thereby "capping" the mRNA.
The initiation of the translational process requires a ribosomal binding site, where the small subunit of the ribosome binds to the mRNA to form the initiation complex. The first triplet of nucleotides to be translated is the start codon, which usually has the sequence AUG. In prokaryotes, a Shine-Dalgarno sequence is positioned 4 to 13 nucleotides upstream of the start codon. This purine-rich sequence base-pairs with the 3′-end of the 16 S ribosomal RNA (rRNA) and thereby regulates the specificity of ribosomal binding. So-called specialized ribosome systems can be constructed that direct ribosomes specifically to the mRNA of interest. Site-directed mutagenesis is applied to both the Shine-Dalgarno region and the rRNA to change their sequence, while maintaining the base pairing. In eukaryotes, the formation of a translational initiation complex is directed by the 5′-cap -structure, 7MeG(5′-5′)pppNp (N is the first nucleotide of the mRNA), at the 5′-end of the mRNA. However, several eukaryotic mRNAs have been found to be translated in a cap-independent manner (2).
Secondary structures in the 5′-untranslated region of the mRNA and around the start codon may hamper recognition of the start site. If potential secondary structures seem to be any problem, they can be eliminated by introduction of silent mutations at the third positions of the initial codons or by random mutagenesis in the region upstream of the start codon. Another approach to overcome the problem of secondary structures is to express the protein of interest as a fusion protein or to use a two- cistron system. In the first approach, a nucleotide sequence encoding a protein or peptide that is known to be well expressed is introduced in-frame between the start codon and the gene of interest. The resulting N-terminal fusion partner most often provides other advantages as well, such as increased stability or simplified purification (see Fusion Gene, Fusion Protein). In the second approach, a strong ribosomal binding site and the 5′-end of a well-translated gene, followed by a stop codon, is positioned immediately upstream of the gene of interest. Translation of the protein of interest will then be reinitiated immediately after the first gene.
The degeneracy of the genetic code implies that most of the 20 amino acids are encoded by two or more codons called synonymous codons. The synonymous codons are not used with equivalent frequencies by different strains or even throughout the genome of a single organism (3, 4) (see Codon Usage and Bias). Weakly expressed genes are characterized by the occurrence of infrequently used codons, which are typically recognized by rare tRNA species. It is advisable to avoid the use of rare codons when possible in systems for high-level expression.
The three stop codons UAG, UAA, and UGA differ in their efficiency in terminating translation. UAA seems to be favored in highly expressed genes and [should be used in] expression systems.
If a heterologous expression system is used, the gene of interest should preferably be devoid of introns, which may not be properly processed by a host of different origin than the gene. However, the presence of introns has, in some cases, been required for successful expression. In such cases, the intron can be provided by the expression vector downstream of the gene of interest.
2. Choice of host
Considerable knowledge about expression in Escherichia coli has accumulated over the years, and E. coli is often the first choice of host when a new protein has to be expressed. Furthermore, E. coli grows fast in inexpensive media, allowing scale-up for industrial purposes. However, proteins containing disulfide bonds are unlikely to fold correctly in the cytosol of bacteria, which is relatively reducing. Misfolding may lead to the formation of inclusion bodies, which can be both an advantage and a disadvantage. The advantages are that the protein is protected from proteolytic degradation and that the purification strategy can be simplified. However, the protein needs to be denatured and refolded, a task that can cause difficulties (see Protein Folding In Vitro). The coexpression of folding factors such as molecular chaperones, thioredoxin, or protein disulfide isomerase, which assist the folding of protein, can be beneficial in some cases. A secretion strategy may represent another solution to the problem [see Secretion Vector]. Even though E. coli is able to secrete proteins, the outcome of such an approach is highly unpredictable. Bacillus species will often be a better choice of host in such a case. Furthermore, Bacillus has the advantage that it does not produce endotoxins (lipopolysaccharides) and has been classified as a GRAS (which stands for generally regarded as safe) organism. The range of promoters available for Bacillus is more limited than the range for E. coli. Some other prokaryotic hosts, eg, Lactococcus lactis (5), also are gaining popularity as expression hosts.
On the other hand, bacterial systems have this limitation: they are unable to provide many of the post-translational modifications often found in eukaryotic proteins. If these modifications are needed for obtaining proper structure or activity, a eukaryotic host should be chosen.
Yeast, the simplest eukaryotic expression host, offers many advantages of both prokaryotic and eukaryotic systems. Yeasts grow rapidly in inexpensive media and are easy to manipulate genetically. Furthermore, yeasts provide an environment for carrying out secretion and providing post-translational modifications that are more similar to those found in proteins from the higher eukaryotes. The yeast strain traditionally used as a host for protein expression, Saccharomyces cerevisiae, is regarded as safe, given its long history of use in the production of food and beverages. More recently, the methylotrophic yeasts Pichiapastoris (6) and Hansenulapolymorpha have gained popularity because of their higher production levels. However, yeast shows a tendency to hyperglycosylate the overexpressed protein, and the resulting high-mannose polysaccharide structures may affect the activity or folding or both, and they are potentially immunogenic. The filamentous fungi, Aspergillus and Trichoderma, are becoming increasingly popular as expression hosts, not least because of the very high amounts of protein that can be obtained with secretion systems (7).
Another eukaryotic expression system of high popularity is the baculovirus/insect cell system, which is relatively easy, cheap, and fast to use. The first generation of systems utilized the strong transcription signals for expression of the polyhedron protein of the virus . The baculovirus system provides more, although not all, of the features characteristic of mammalian cells, and the yields are often high. However, the high yields sometimes lead to the formation of inclusion bodies. Interestingly, the baculovirus system can be used for phage display of large, complex disulfide-containing proteins, thereby overcoming the limitations of the original bacterial system.
If the protein of interest is of mammalian origin and if authenticity is of utmost importance, a mammalian expression system should be chosen. It may also enable the study of an engineered protein in its natural environment; for example, a biological assay could be coupled to the expression system for the evaluation of functional effects. The basic technology is now available, but these expression systems are not very cost-efficient and are sometimes difficult to scale up. Often, it takes a long time to establish a stable system with high expression levels.
Eukaryotic expression systems can be divided into two groups: those that involve transient or stable expression of recombinant genes from transfected DNA molecules, and those that involve helper-independent viral expression vectors. Vectors used for stable expression contain a complete eukaryotic transcriptional unit inserted into a bacterial replicon. The DNA integrates at a low frequency into the host genome and usually directs the expression of the desired protein at low levels. Recombinant virus systems represent powerful tools for the expression of recombinant proteins in cultured cells, animals and man. A comparison of five different eukaryotic expression systems can be found in reference 8.
There are several factors to take into consideration before setting up an expression system. How much protein is needed? What is its application? Is authenticity important? Can the presence of heterogeneity be accepted? Are the technologies available? The choice of an expression system will depend largely on the desired use of the expressed protein; for example, even microheterogeneity can be a problem if the protein is to be used for X-ray crystallographic structural analysis, as these heterogeneities may hamper the crystallization process (9).
Proteolysis of heterologous proteins is another problem that should be considered in the choice of a host for expression. Bacteria and lower eukaryotes use proteolysis as a primitive immune system, which attacks and eliminates "nonself." Different proteinase-deficient strains have been constructed to overcome this problem, but these strains grow at a lower rate, and their usefulness for solving a particular problem is often unpredictable. If proteolysis is a problem, it is advisable to test a selection of these hosts. Alternatively, a secretion strategy or a fusion protein strategy can be chosen. In the first case, the gene product is removed from the cytoplasm, where most proteinases are located. In the second case, the fusion partner can have a stabilizing effect. The method of induction of expression should also be considered when proteolysis appears to be a problem. Some induction methods (eg, heat induction) activate the heat-shock system of the cell, which includes a whole range of proteolytic enzymes.
In particularly difficult cases, where the protein of interest is toxic or prone to form inclusion bodies, the use of a cell-free protein biosynthesis system may be beneficial. In such a system, the enzymes needed for transcription and translation are present in a cell extract instead of a live organism. One further advantage is the possibility of introducing unnatural amino acids into the protein of interest (10, 11).
Finally, when a suitable expression system has been established, the growth and eventual induction conditions need to be optimized to obtain the maximal yield of product. It is often advisable to decrease the growth rate below the optimal by reducing the temperature, aeration, or nutrition content of the media. The risk of overloading the protein synthesizing machinery, leading to inclusion body formation or misfolded proteins, is thereby avoided. Furthermore, the expression vector is stabilized. Also, the strategy of recovering the expressed protein will influence the product yield and thus deserves optimization.
The vast number of vectors and hosts available should be able to satisfy the demands of any protein to be expressed. Yet protein expression is a somewhat empirical process, making it difficult to foresee which system should be chosen for optimal success. Advances in the understanding of how expression systems work will undoubtedly make for a higher degree of predictability than is currently the case.