One of the fundamental methods of molecular biology is determining the sequences of bases in specific segments of DNA. The sequence information can be used to help deduce the function of the DNA segment, map its chromosomal location, determine how it might be involved in regulating gene expression or replication, or elucidate how it interacts with proteins. For example, the sequence of an unknown complementary DNA (cDNA) can be used to deduce the sequence of the protein encoded by its parent messenger RNA. In turn, this protein sequence can be compared with the sequences of all known proteins, giving clues to function and evolutionary origin (see Sequence Analysis).
1. Chain Termination, Sanger Method
The chain-termination method of DNA sequencing was first described by Sanger in 1977. This method involves synthesizing a DNA strand by a DNA polymerase in vitro. Synthesis is initiated at only one site, where a primer anneals to the template. The growing chain is terminated by incorporating a 2′,3′-dideoxynucleoside triphosphate (ddNTP) that does not support continued DNA synthesis (hence the name chain termination).
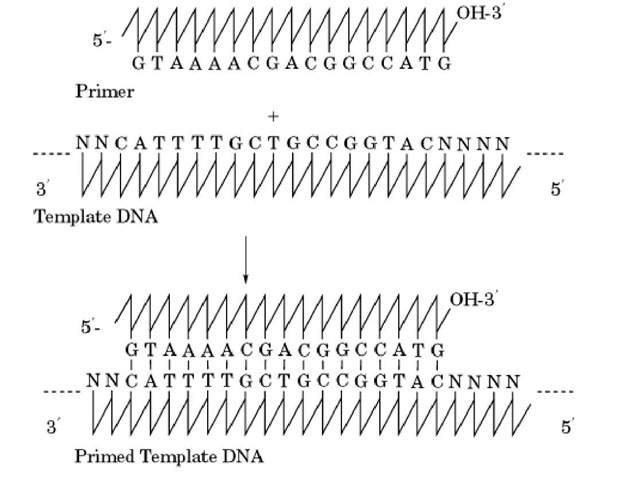
DNA polymerases initiate synthesis only at the 3′-end of a primer annealed to a DNA template. For most sequencing applications, the primer is a short synthetic oligonucleotide (18 to 35 nucleotides long) that is complementary in sequence to the template at a unique position adjacent to the region to be sequenced. The primer is hybridized to the template at the appropriate temperature (Fig. 1). Once this duplex is formed, the primer is extended by the DNA polymerase in the presence of the four deoxynucleoside triphosphates (dGTP, dATP, dTTP, and dCTP; dNTP corresponds to any one of the four).
Figure 1. Annealing of primer to template DNA.
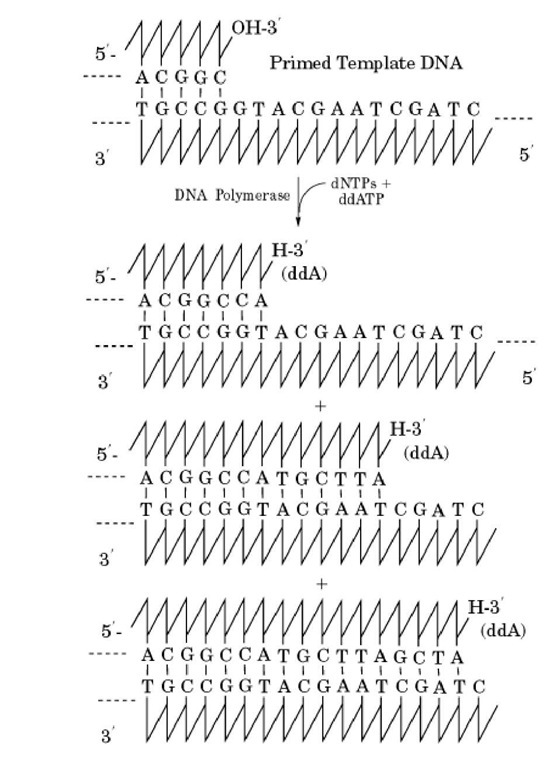
Polymerization of the new strand requires a free 3′-hydroxyl group (Fig. 2). As long as dNTPs are incorporated, there is a 3′ hydroxyl group available for continued polymerization of the growing chain (Fig. 3). However, dideoxynucleoside triphosphates (ddNTPs) lack a 3′-hydroxyl group and terminate chain elongation when incorporated into the DNA. Consequently, synthesis, directed by a sample of template DNA that has a unique primer, in the presence of all four deoxy- and one dideoxynucleoside triphosphate yields a population of molecules that have a common 5′-end, plus 3′-ends all of which have the same terminal dideoxynucleotide base, but have a distribution of sizes depending on the site at which the ddNTP is incorporated (Fig. 4). Thus, the size of each fragment is determined by the sequence of the template. Typically, four separate reactions are performed, each with a different ddNTP. The products of the four reactions are analyzed by electrophoresis using a denaturing polyacrylamide gel, which accurately separates the products by size. Because the size of each fragment is determined by the template sequence, this sequence can be determined from the order of the bands on the gel.
Figure 2. Biochemistry of chain elongation. In the presence of dNTPs, DNA polymerase catalyzes the condensation of deoxynucleoside triphosphate at the 3′-end of a primed template, releasing pyrophosphate.
Figure 3. Chain elongation by DNA polymerase. As long as dNTPs are incorporated, there is a free 3′-hydroxyl group available for continued polymerization of the growing chain.
Figure 4. Chain termination. Synthesis from a unique primer in the presence of all four deoxy- and one dideoxynucleoside triphosphate yields a population of molecules with common 5′ ends, but different 3′-ends, depending on the site at which a ddNTP is incorporated.
2. Chain Cleavage, Maxam-Gilbert Method
Another general sequencing method, known as the chain-cleavage or Maxam-Gilbert method, has also been used extensively. This method works by a similar method of mapping DNA sequence to DNA size, but does so by degrading existing DNA chains, rather than synthesizing new ones. A sample of purified DNA to be sequenced is first labeled at one end. Then the DNA is subjected to a chemical treatment that breaks each DNA molecule at random, but only at places where one (or a defined subset) of the bases occurs. The result is a population of labeled molecules whose sizes are determined by the sequence. Determination of the sizes on samples cleaved with several sequence-specific cleavage treatments yields complete sequence information. Unfortunately, it is difficult to find chemical cleavage conditions that consistently give unambiguous, clean cleavage products, and few labeling methods can be used that provide labels stable enough for these treatments. For these and other technical reasons, chain-cleavage methods are rarely used today.