1. Introduction
After the discovery of humoral immunity, Hans Buchner found that if fresh serum possessing an antibacterial antibody was added to bacteria, the bacteria were quickly lysed. However, if the serum was heated to 56°C, the lytic capacity of the serum was lost. He also demonstrated that the loss of lytic capacity could be restored by using unheated nonimmune serum in conjunction with heated immune serum. These studies were extended by others, including Jules Bordet, who concluded that serum contained two components necessary for cellular lysis. These two components are heat-stable antibodies and a heat-labile component that "complements" the lytic function of antibodies. He reasoned that antibodies had two binding sites, one for antigen and the other for heat-labile substance that was given the name complement.
Complement is now known not to be a single component, but it consists of at least 20 chemically and immunologically distinct plasma proteins capable of interacting with one another in a highly regulated manner to provide at least four main biological functions. First, cytolysis is mediated by the association, or polymerization, of specifically activated complement components on the surface of targets cells. These components form pores that disrupt the integrity of the lipid membrane, and the cell is killed by osmotic lysis. Second, antibody and antigen combine to form immune complexes that, unless removed, can result in damage of body tissues. The binding of complement proteins prevent the damage from immune complexes by mediating their solubilization and clearance. Third, the opsinization of foreign particles is mediated by the binding of complement proteins (opsonins). Phagocytic leukocytes bear receptors for these complement proteins, so that opsinized particles are cleared from the body by phagocytosis. Finally, through the activation of complement, proteolytic fragments are released whose function are to mediate inflammation. These fragments, or "anaphylatoxins," can act on several target cells such as neutrophils, smooth muscle, and vascular endothelium, as well as organ systems of the body.
The complement proteins are normally present in the circulation and are produced primarily by the liver and other extrahepatic sources, such as macrophages and fibroblasts. Some of the components are produced as functionally inactive proteinases that are activated only when proteolytically cleaved themselves by previously activated complement proteins. Complement activation occurs only at localized sites under specific conditions. The binding of specific antibody to antigen can initiate complement activation through what is called the classical pathway. Additionally, in the absence of antibody, some complement components are directly activated by binding to the surfaces of infectious agents, such as bacteria, fungi, and viruses, through what is called the alternative pathway.
2. Overview of the Complement System
Individual proteins of the complement system are present in the serum as functionally inactive molecules. The individual precursor proteins are designated numerically, such as C1, C2, up to C9. Other proteins involved with the complement system have retained their common, or trivial, names such as properdin, factor B, and factor H. Individual components must be activated sequentially and specifically for the complement cascade to progress and mediate inflammation and lysis of foreign organisms. The activation of complement is a dynamic process that allows the proteins of the system to interact functionally and is not a static singular event.
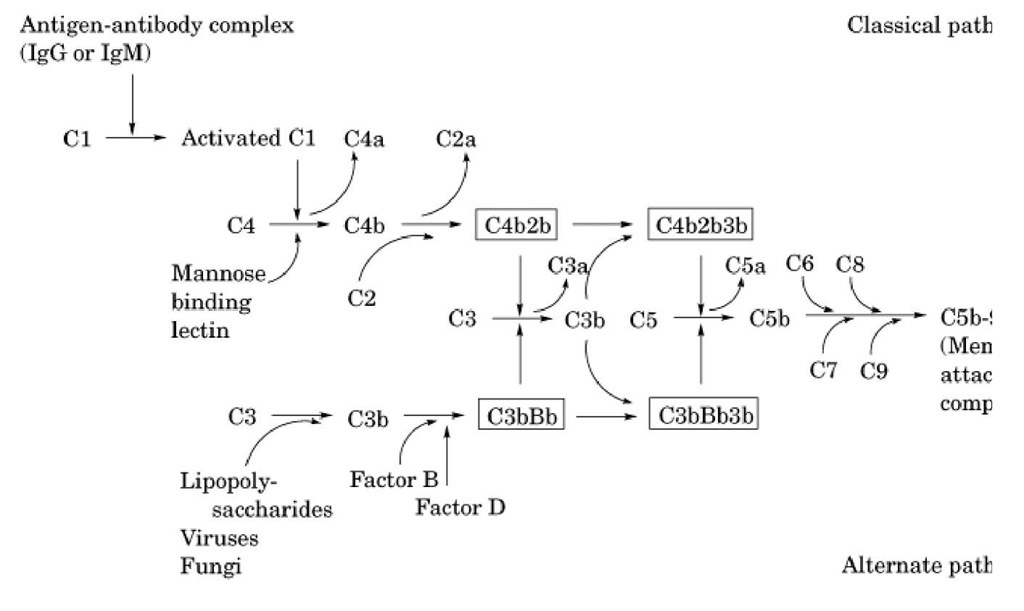
An overview of the operation of the complement system is shown in Figure 1, and biochemical and physical information of the proteins involved in the complement system are listed in Table 1. A protein called C3 is the central component of the complement system. The alternative and classical pathways are two parallel, but entirely independent, mechanisms that lead to the formation of C3 convertases whose function is to cleave C3 into C3a and C3b. In the classical pathway, initial events involve binding of specific antibody to antigen. This antigen-antibody complex binds complement C1 and sequentially activates complement proteins C4 and C2, leading to the formation of the C4b2b complex that functions as the classical pathway C3 convertase. In contrast, activation of complement via the alternative pathway occurs independent of antigen-antibody complexes. Instead, alternative pathway activation is dependent on activators that allow the formation and deposition of the C3bBb complex (alternative pathway C3 convertase) on their surfaces. Activators of the alternative pathway include many bacteria, fungi, and viruses. Both classical and alternative convertases can cleave additional C3, generating more C3b (see Fig. 1). The binding of additional C3b to these C3 convertases changes them conformationally to C5 convertases, which specifically cleave C5. Once cleaved, both the alternative and classical pathways share the same terminal steps. These steps do not involve the proteolytic cleavage of additional components but instead involve the sequential binding of components C6, C7, C8, and C9. This terminal assembly leads to the formation of the membrane attack complex (MAC), resulting in the osmotic lysis or cytolysis of bacteria or other affected cells.
Figure 1. Schematic representation of the alternative and classical activation of the complement pathway leading to the assembly of the membrane attack complex.
Table 1. Protein Components of the Classical and Alternative Pathwaysa
|
Serum |
Number of mRNA- |
Gene6 |
||||
|
Concentration, Molecular Polypeptide |
Size, |
Chromosome |
Size, |
|||
|
Component |
^g/mL |
Weight |
Chains |
kb |
Location |
kb |
|
Classical |
Pathway |
|||||
|
C1q |
75 |
410,000 |
6 of A at |
0.52 |
1 |
2.5 |
|
24,000 |
||||||
|
6 of B at |
1 |
1 |
2.6 |
|||
|
23,000 |
||||||
|
6 of C at |
1.6 |
1 |
3.2 |
|||
|
22,000 |
||||||
|
C1r |
34 |
85,000 |
Single chain |
2 |
12 |
10.5£ |
|
C1s |
30 |
85,000 |
Single chain |
2 |
12 |
10.5£ |
|
C4 |
450 |
210,000 |
a-Chain, |
5.3 |
6 |
16 |
|
93,000; b- |
||||||
|
chain, |
||||||
|
75,000; g- |
||||||
|
chain, |
||||||
|
33,000 |
||||||
|
C2 |
25 |
95,000 |
Single chain |
2.9 |
6 |
18 |
|
C3 |
1500 |
195,000 |
a-Chain, |
5.2 |
19 |
41 |
|
110,000; b- |
||||||
|
chain, |
||||||
|
75,000 |
||||||
|
C5 |
75 |
180,000 |
a-Chain, 115,000; b-chain, 75,000 |
5.5 |
9 |
80 |
|
C6 |
60 |
128,000 |
Single chain |
5 |
85 |
|
|
C7 |
60 |
121,000 |
Single chain |
3.9 |
5 |
78 |
|
C8 |
80 |
150,000 |
a-Chain, 64,000; |
2.5 |
1 |
70 |
|
b-chain, |
2.6 |
1 |
40 |
|||
|
68,000; |
||||||
|
g-chain, |
1 |
9 |
1.8 |
|||
|
22,000 |
||||||
|
C9 |
58 |
79,000 |
Single chain |
2.4 |
5 |
80 |
|
Mannose |
1 |
~ 600,000 |
Multimer of |
3.5 |
10 |
7 |
|
binding Lectin |
32,000 |
|||||
|
Alternative Pathway |
||||||
|
Properdin |
25 |
220,000 |
4 at 56,000 |
1.6 |
X |
6.4 |
|
Factor B |
225 |
100,000 |
Single chain |
2.9 |
6 |
6 |
|
Factor D |
1 |
25,000 Inhibitors |
Single chain —Soluble |
1 |
4 |
2.5d |
|
Factor I |
34 |
105,000 |
46,000 |
2.4 |
4 |
63 |
|
Factor H |
500 |
150,000 |
Single chain |
4.4 |
1 |
7 |
|
C1 inhibitor |
275 |
105,000 |
Single chain |
1.8 |
11 |
17 |
|
C4 binding |
150 |
560,000 |
6, a-Chain, |
2.5 |
1 |
40 |
|
protein |
70,000; 1, |
|||||
|
b-chain, |
1 |
1 |
10 |
|||
|
45,000 |
||||||
|
S protein |
500 |
83,000 |
Single chain |
1.6 |
17 |
5.3 |
|
Membrane—Inhibitors and Receptors |
||||||
|
DAF (CD55) |
70,000 |
Single chain |
3.1 |
1 |
40 |
|
|
MCP (CD46) |
58,00063,000 |
Single chain |
4.2 |
1 |
43 |
|
|
CR1 (CD35) |
190,000280,000 |
Single chain |
7.3-13 |
1 |
130160 |
|
|
CR2 (CD21) |
140,000 |
Single chain |
5 |
1 |
20 |
|
|
CR3 |
260,000 |
a-Chain, |
4.1 |
16 |
55 |
|
|
(CD11b/CD18) |
165,000; |
|||||
|
b-chain, |
3 |
21 |
32 |
|||
|
95,000 |
||||||
|
CR4 |
245,000 |
a-Chain, |
4.7 |
16 |
25 |
|
|
(CD11c/CD18) |
150,000; |
|||||
|
b-chain, |
3 |
21 |
32 |
|||
|
95,000 |
||||||
|
MIRL (CD59) |
18,000 |
Single chain |
0.6-6.0 |
1 |
27 |
|
|
C5aR (CD88) |
42,000 |
Single |
3.0 |
19 |
9 |
|
|
chain2 |
||||
|
C3aR |
50,000 Single |
3.0 |
12 |
8 |
|
chain2 |
a All data for proteins listed are for human complement components. b Approximate size.
c C1r size estimated from proximity and homology to C1s. d Complete factor D cDNA hybridizes within a 2.5-kb genomic fragment. e Single chain with 7-transmembrane domains.
During activation, through either the classical or alternative pathway, various cleavage products mediate a variety of biological functions. As discussed, cytolysis is mediated through the membrane attack complex. Opsinization and subsequent phagocytosis by leukocytes is mediated primarily through C3b, a cleavage fragment of C3 that binds receptors for C3b. Cleavage products of C3, C4, and C5 (C3a, C4a, and C5a anaphylatoxins) mediate inflammatory events and the recruitment and activation of leukocytes.